Coded the regression analysis
on Thursday, September 7th, 2023 2:16 | by Björn Brembs
I now switched the sign of the Optomotor Asymmetry Index in flies that were punished on producing right-turning torque, such that weaker punished torque shows up as a positive index. After that was done, I plotted the correlation between the optomotor index and the preference index:
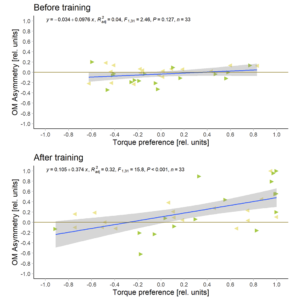
I had to get rid of eight flies where the optomotor response was already asymmetric before the training started, so now I only have 33 flies. But with these flies, there is no correlation before training and a very significant correlation after training.
Any suggestions about appearance of the graphs?
Would it be useful to plot optomotor and performance indices as raincloudplots next to the regressions?
This would be the complete figure:
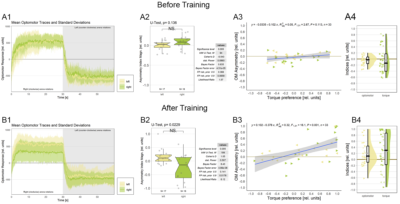
Category: operant self-learning, Optomotor response | No Comments
Optomotor project nearing completion
on Monday, September 4th, 2023 1:59 | by Björn Brembs
The results of comparing optomotor responses after self-learning remain solid. There still is a small asymmetry between the left/right groups, but nothing dramatic:
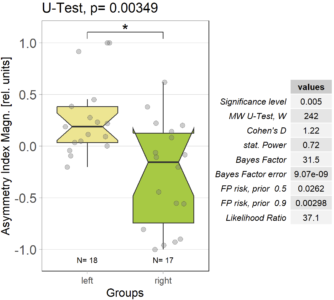
Category: operant self-learning, Optomotor response | No Comments
Optogenetic stimulation of DANs
on Saturday, September 2nd, 2023 11:09 | by Maja Achatz
With the focus on dopaminergic neurons, I conducted two experiments with two different driver lines to see how the naive gustatory behavior in Drosophila larvae is affected.
The first cross I tested was R58E02-Gal4 x UAS-ChR2-XXL, which gave me the following results:
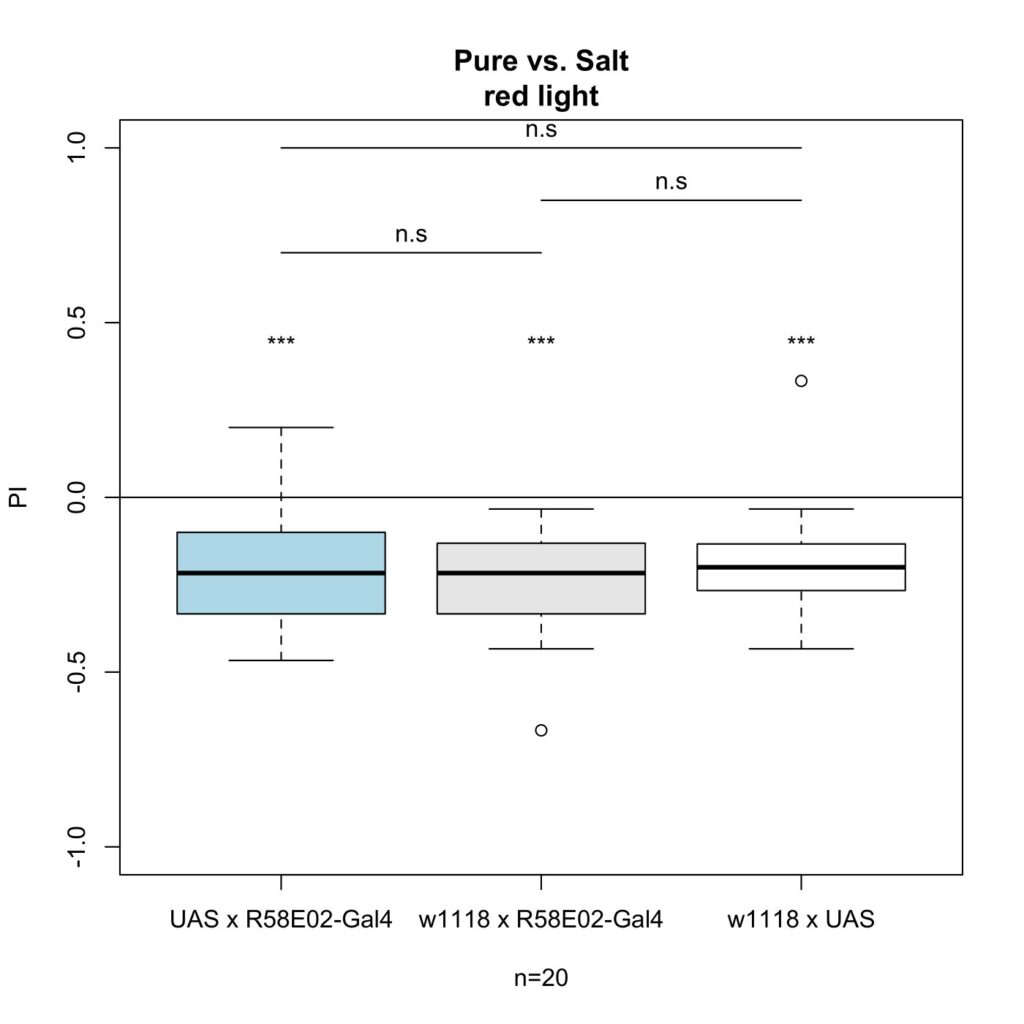
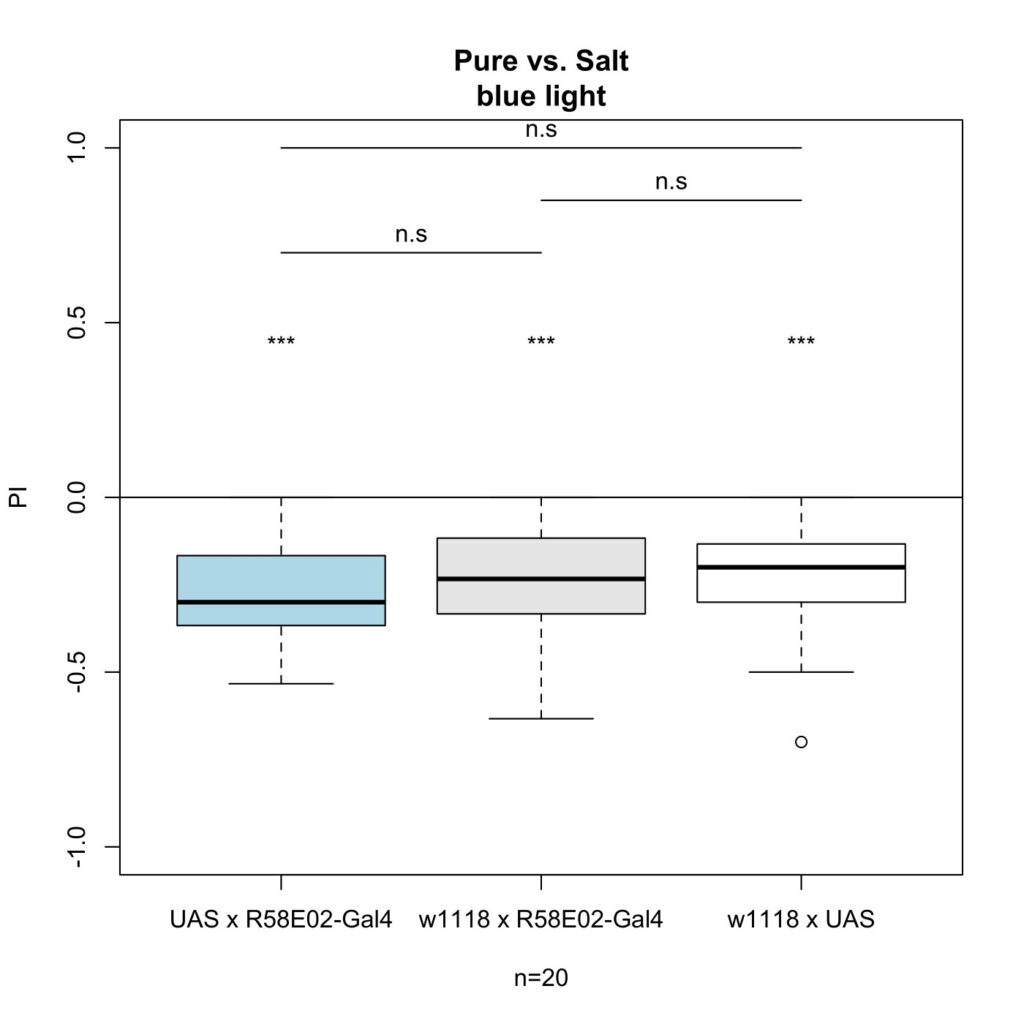
As seen in the figures, there was no effect under blue light.
For the second experiment I crossed TH-Gal4 x UAS-ChR2-XXL with these results:
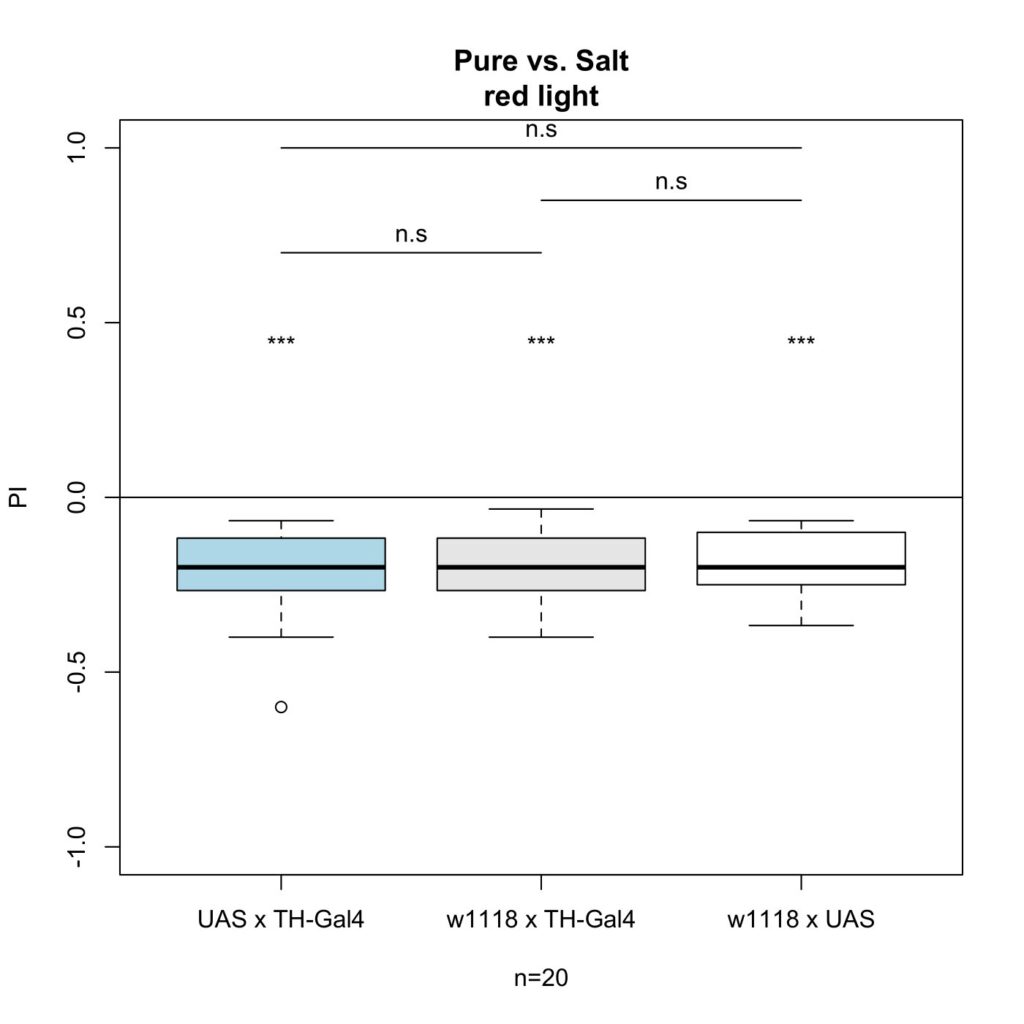
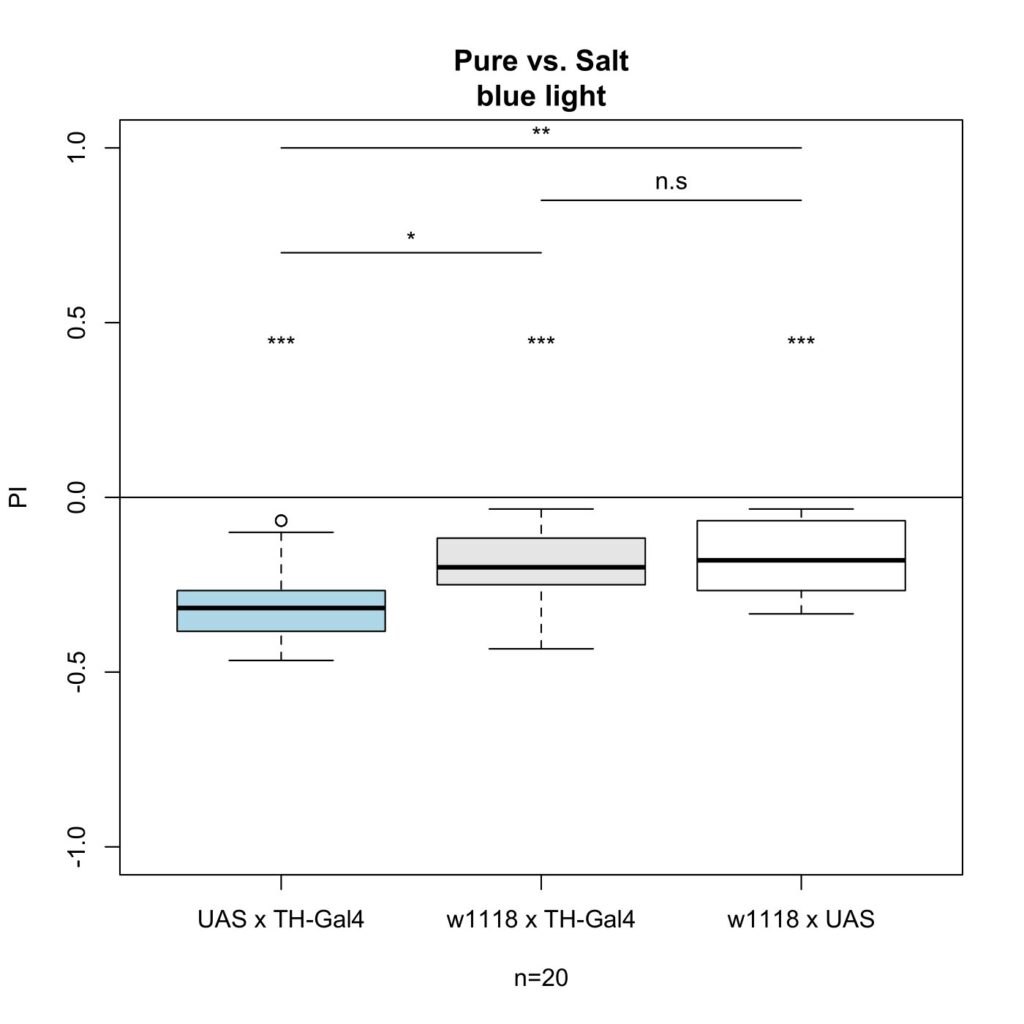
With this experiment, there was an effect on gustatory behavior as there is a clear significant difference between the experimental group and the control group.
Category: DAN, Larve, Optogenetics, PAM | No Comments
Cloning via DNA Assembly
on Friday, September 1st, 2023 7:26 | by Isabel Stark
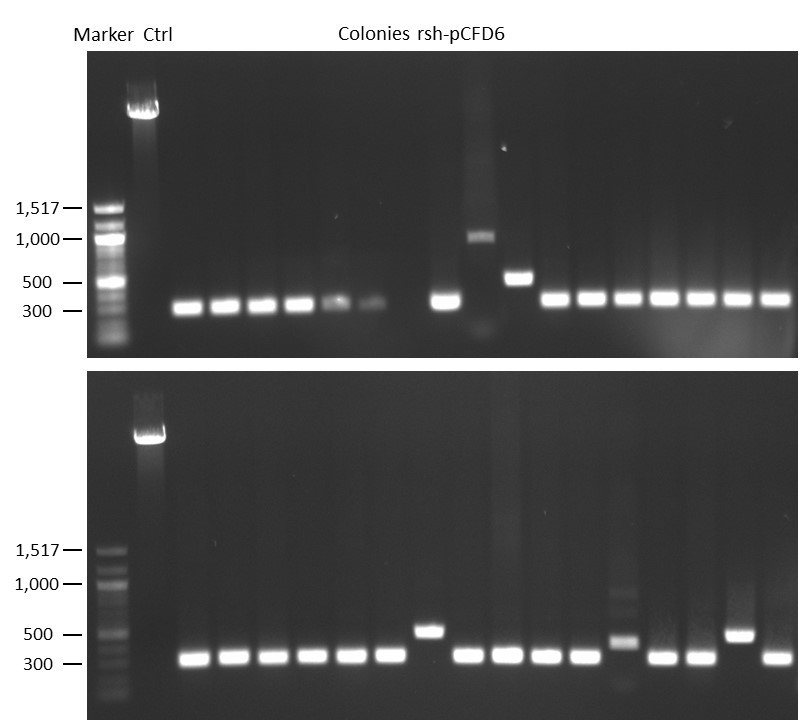
DNA Assembly in a 1:2 ratio of vector to insert with gRNAs of rsh and rut (Q5 and template concentration: 640 pg/µl) and 100ng of pCFD6 BbsI AP (using QuickCIP). Heat-shock (hs) transformation into E. coli (DH5α competent) with 10 µl Assembly Reaction and 100 µl cells.
For Crtl, pCFD6 BbsI AP was wrongly used.
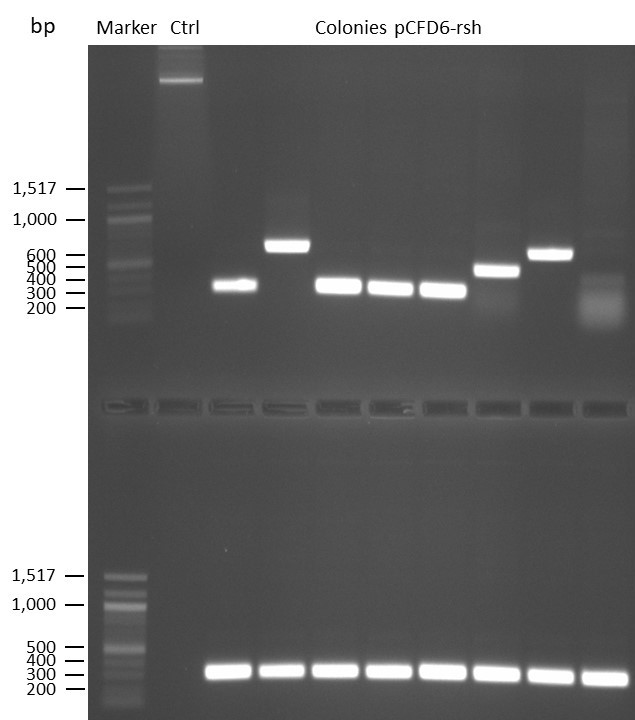
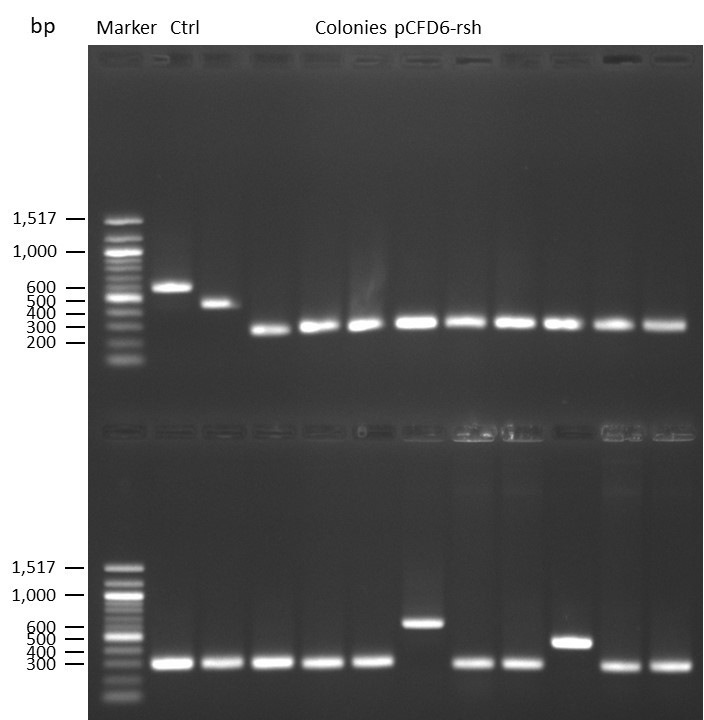
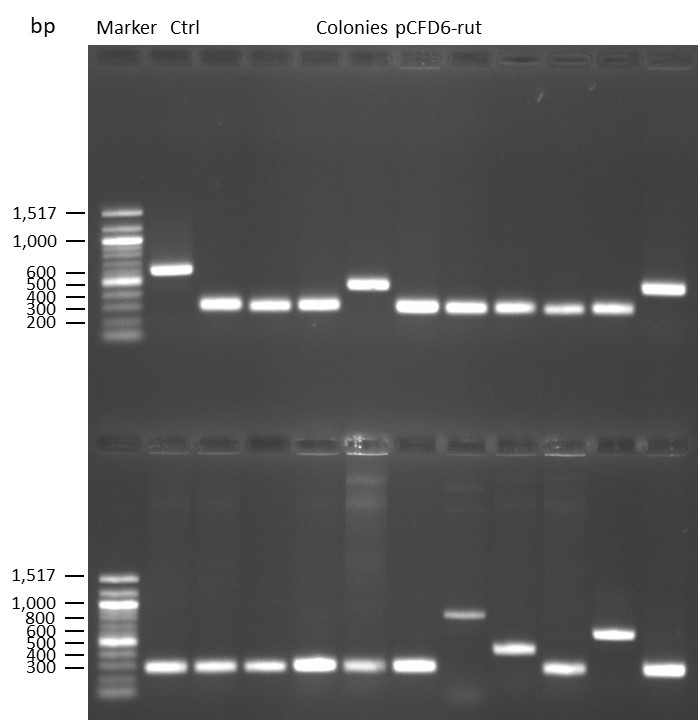
DNA Assembly in a 1:2 ratio of vector to insert with the gRNAs of rsh and rut (Q5 and template concentration: 640 pg/µl) and 100ng of pCFD6 BbsI AP (using FastAP and 2 extraction steps). Heat-shock (hs) transformation into E. coli (DH5α competent) with 10 µl Assembly Reaction and 100 µl cells.
For Crtl, pCFD6 BbsI AP was used in reaction.
Category: genetics, Memory, Operant learning, operant self-learning, Radish | No Comments
Line verification SS56699
on Tuesday, August 29th, 2023 7:50 | by Luisa Guyton
After dissecting brains from the GAL4 driver line SS56699 with GFP staining and finding no fluorescence, I dissected them again and the immunohistochemical staining showed fluorescence. To check that the correct neurons were stained, I compared the images with the image in the paper by Hulse et al (2021; https://doi.org/10.7554/eLife.66039) . The three PPL1 dopaminergic dorsal fan-shaped body tangential neurons per hemisphere are stained, but there are some additional unidentified neurons (presumably PPM1 neruons) visible.
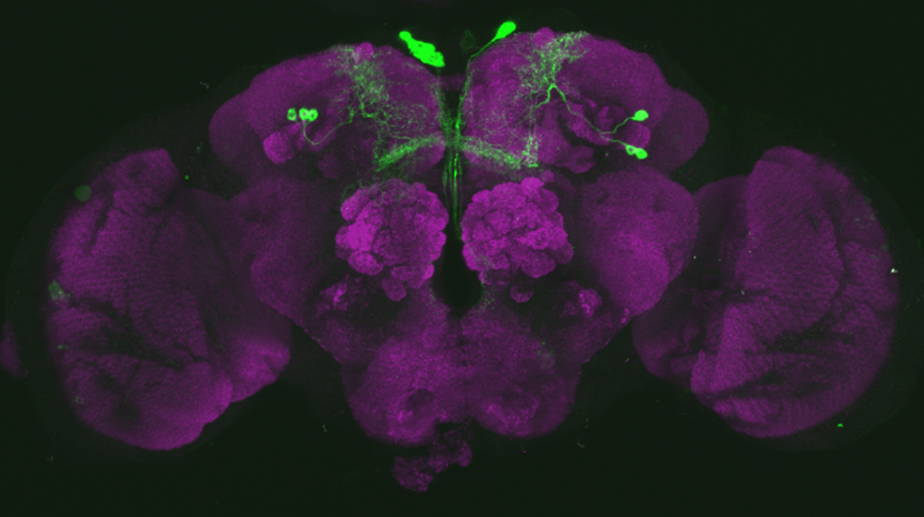
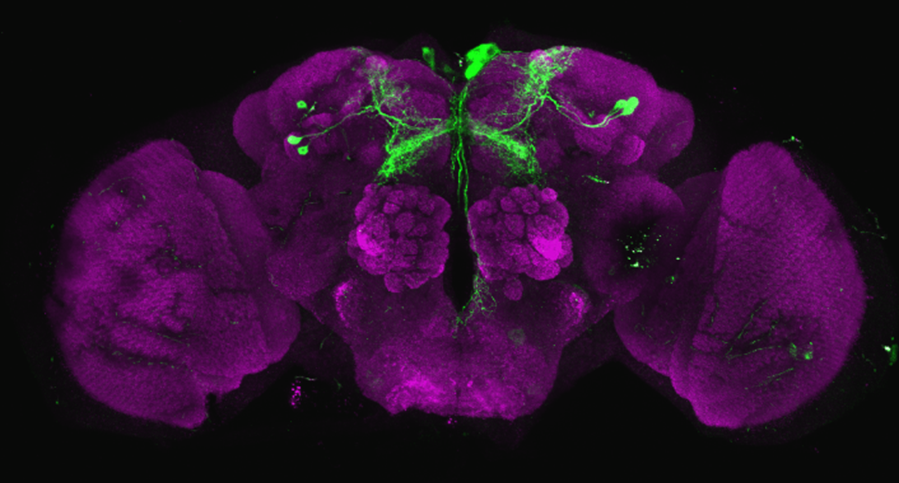
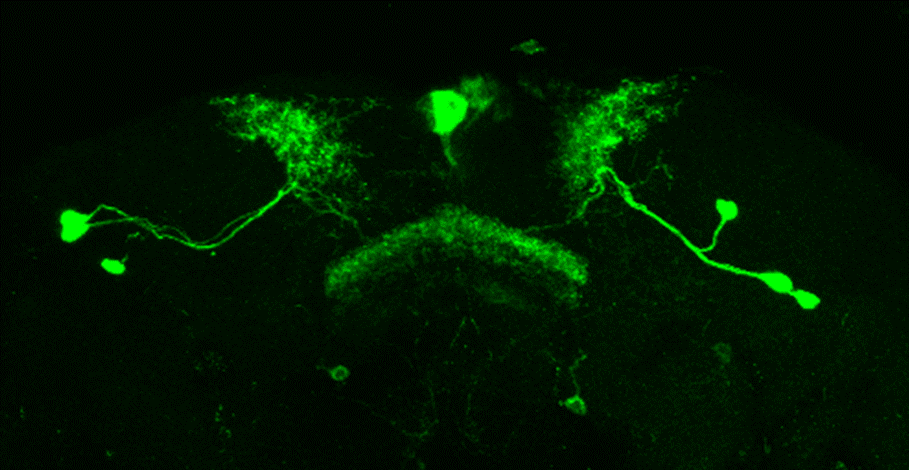
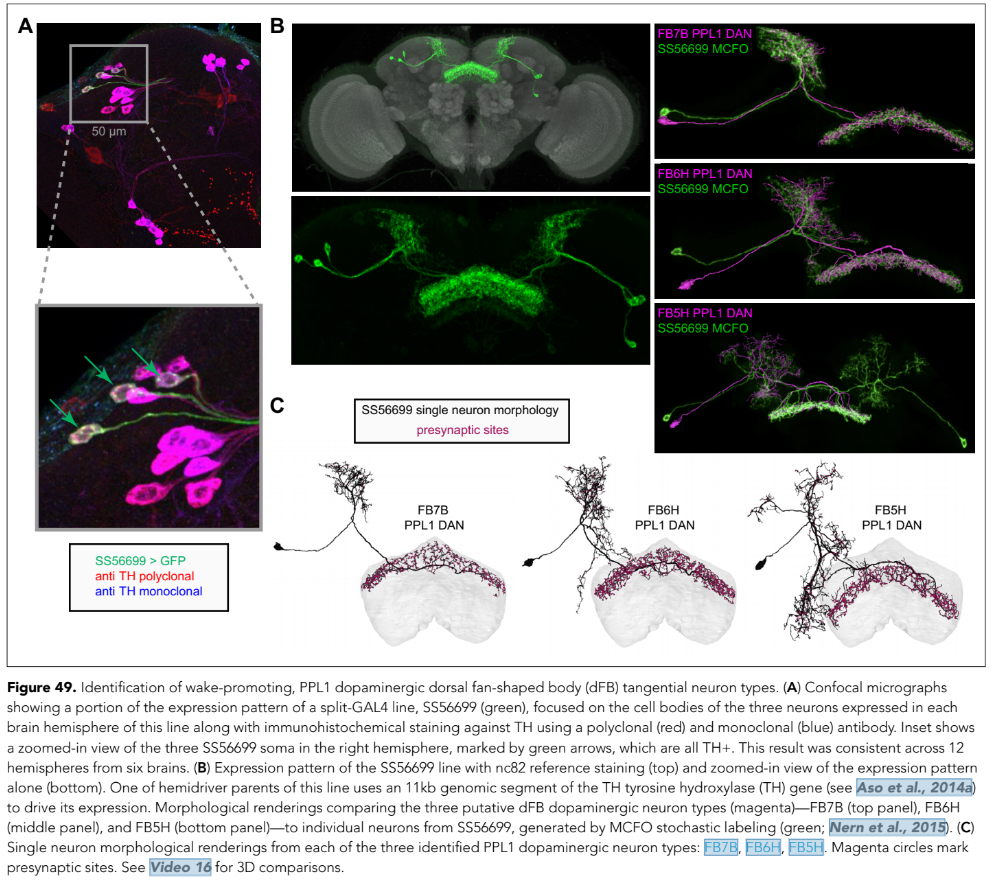
Hulse, B. K., Haberkern, H., Franconville, R., Turner-Evans, D., Takemura, S.-y., Wolff, T., Noorman, M., Dreher, M., Dan, C., Parekh, R., Hermundstad, A. M., Rubin, G. M., & Jayaraman, V. (2021). A connectome of the Drosophila central complex reveals network motifs suitable for flexible navigation and context-dependent action selection. eLife, 10, e66039. https://doi.org/10.7554/eLife.66039
Category: Anatomy, genetics | No Comments
Bachelor Blog / #4 is there something?
on Monday, August 7th, 2023 2:13 | by Ellie
The offspring of my first experimental fly cohort finally hatched! Below you find a few first pre-tests I ran last week :)
First, here are the results of a quick test to see if the offspring shows a preference for the parentally trained side after the first training period:
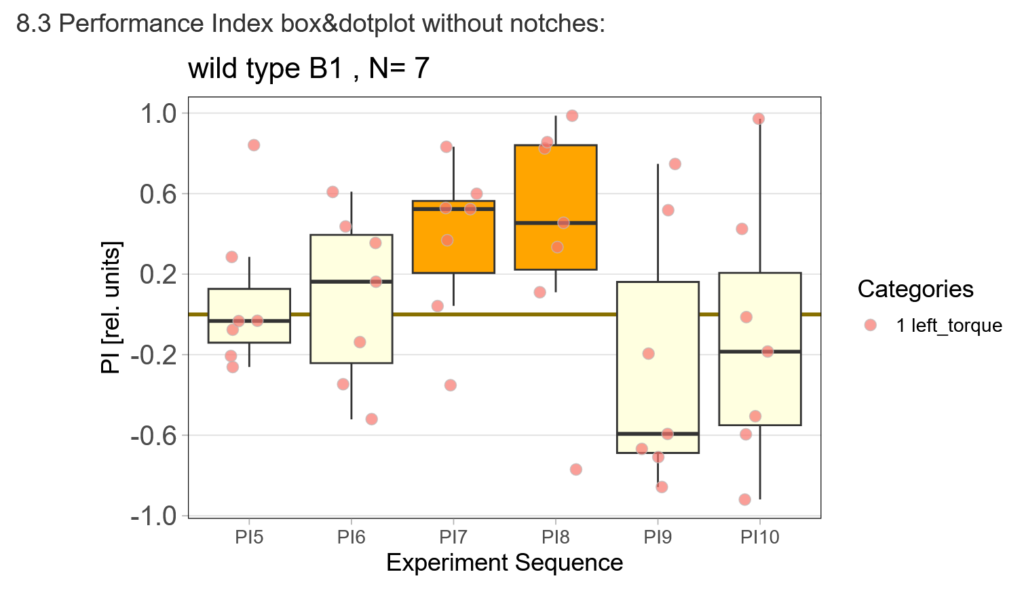
After that I played around with the laser a little bit to find the learning threshhold. I set the laser on 2,6V but the results I got look a bit weird:
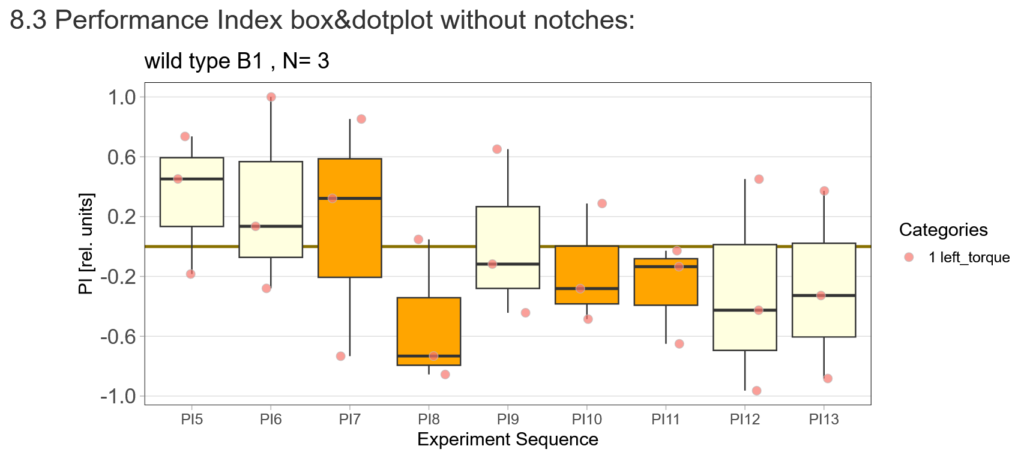
-> untrained wtb flies
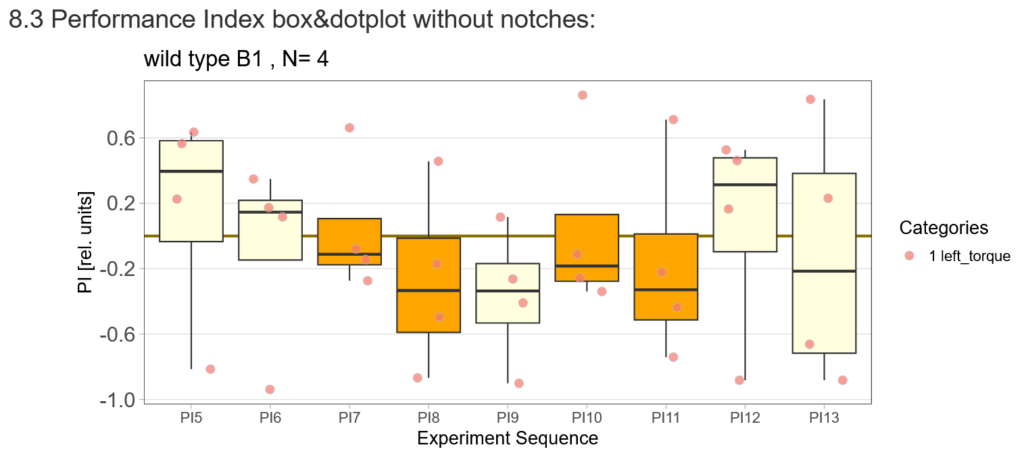
-> offspring of trained wtb flies
I´m optimistic however there is still a lot of work to do…
Preferences with higher salt concentration
on Monday, August 7th, 2023 12:46 | by Maja Achatz
The following plots show the results after I increased the salt concentration from 1,5M to 2,5M. It shows that the difference between the control groups and experimental group are not significant anymore.
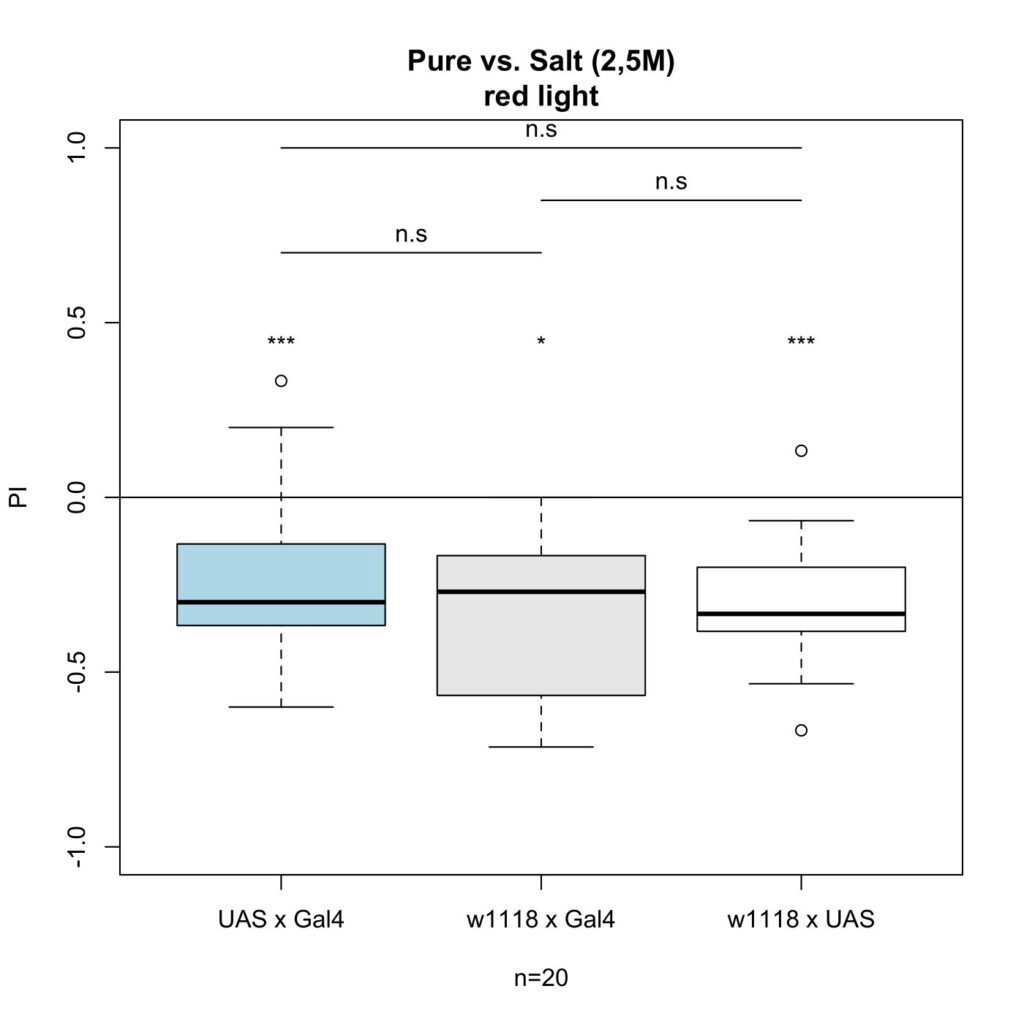
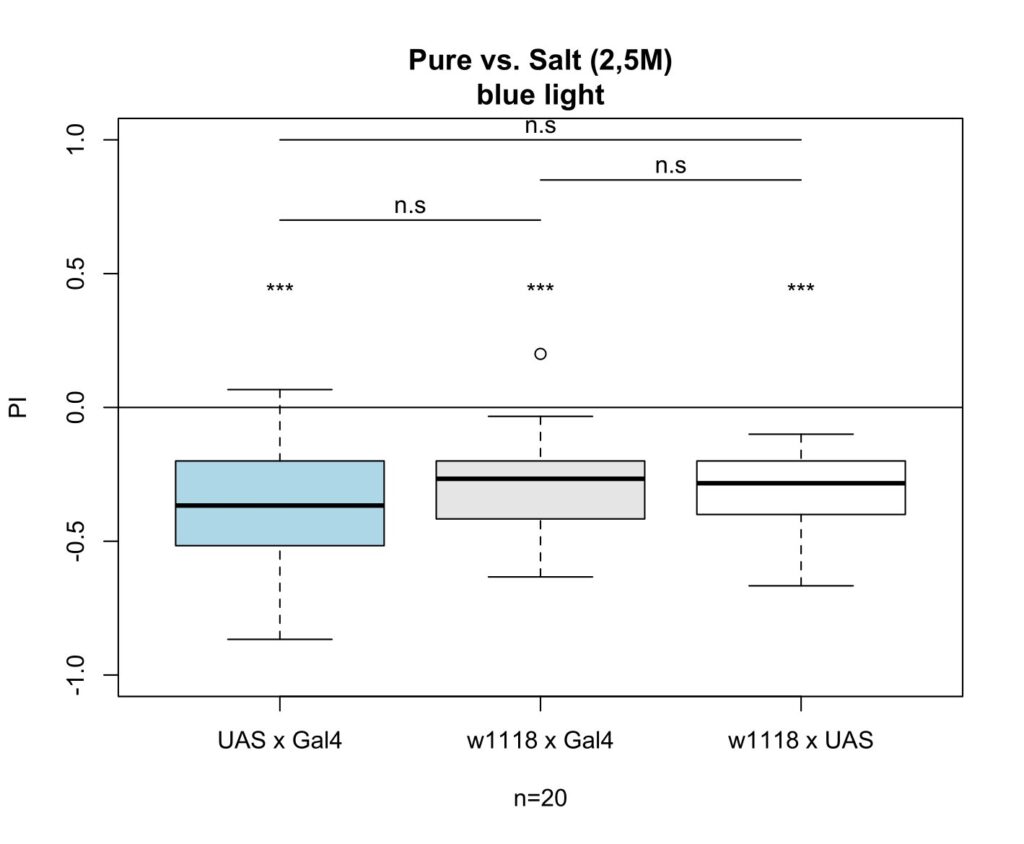
Category: Larve, Mushroom Body, Optogenetics | No Comments
Success: rsh Stock has rsh1 Mutation
on Monday, August 7th, 2023 11:11 | by Isabel Stark
Via gDNA analysis and PCR was the specific area of the rsh gene extracted and amplified where the nucleotide substitution: C to T (Folkers et al., 2006) should be for the rsh1 mutation. The amplicon was Sanger sequenced which proved the nucleotide substitution.
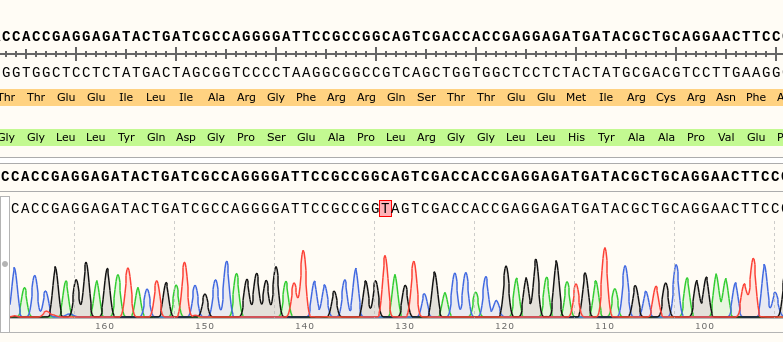
Category: genetics, Memory, Operant learning, operant self-learning, Radish, Uncategorized | No Comments
Little by little
on Monday, July 24th, 2023 1:59 | by Björn Brembs
Despite very warm weather, some flies did fly, even though the learning performance of the control flies was really poor. At least for now, it looks like all stocks are learning and that rut and rsh flies learn at least equally well as the Berlin flies. I’ve also managed to fix the positive preference problem:
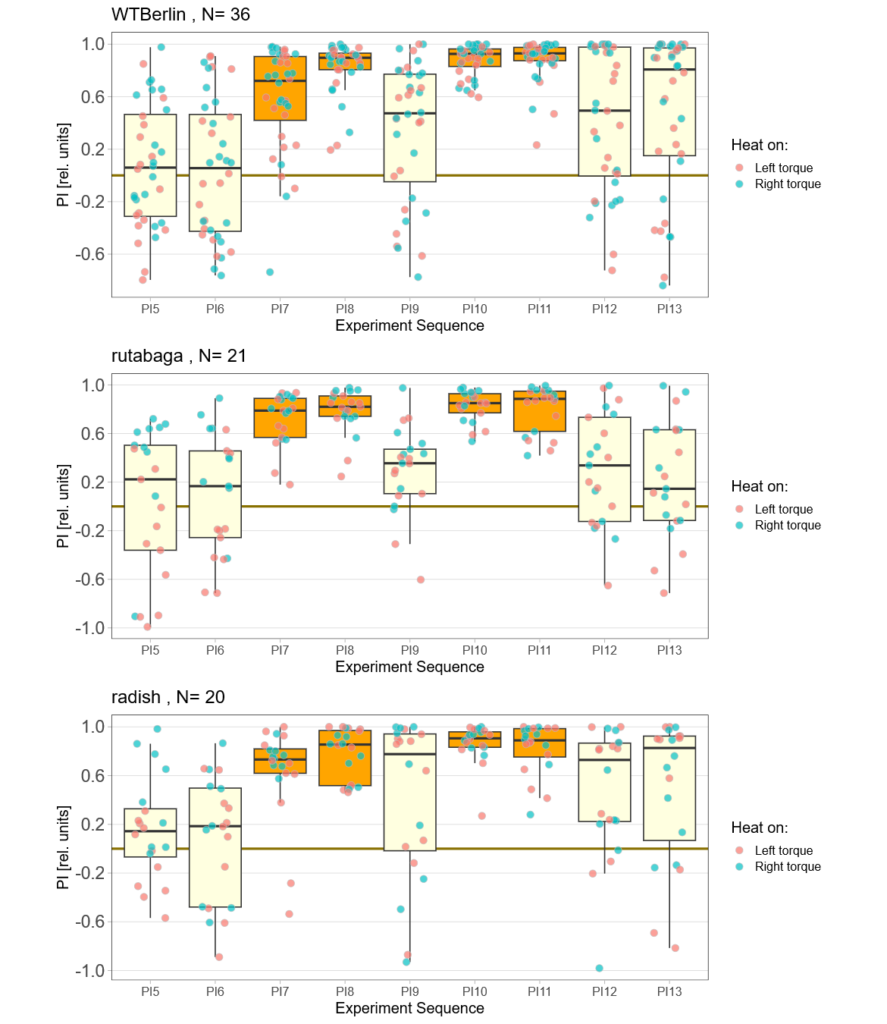
Category: operant self-learning, Radish | No Comments

