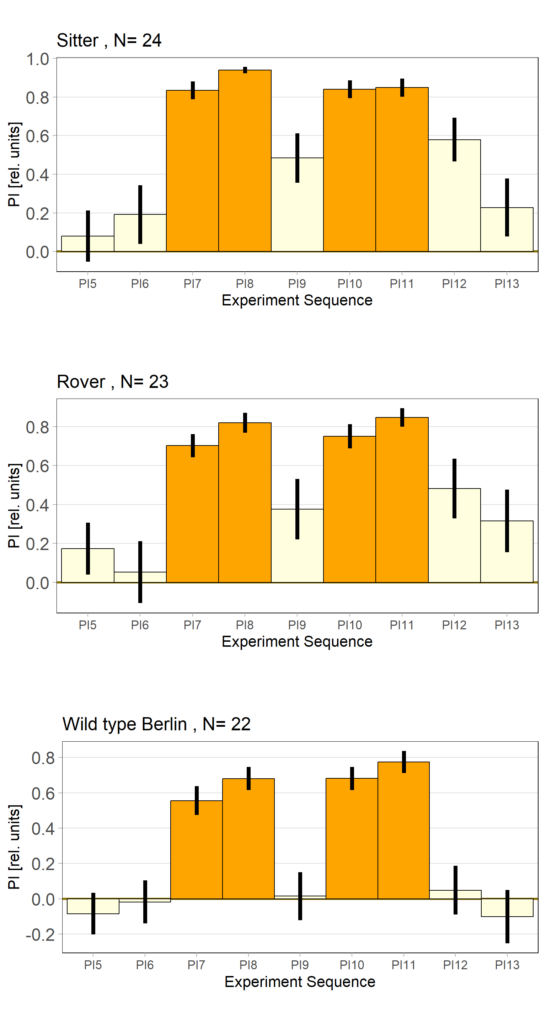
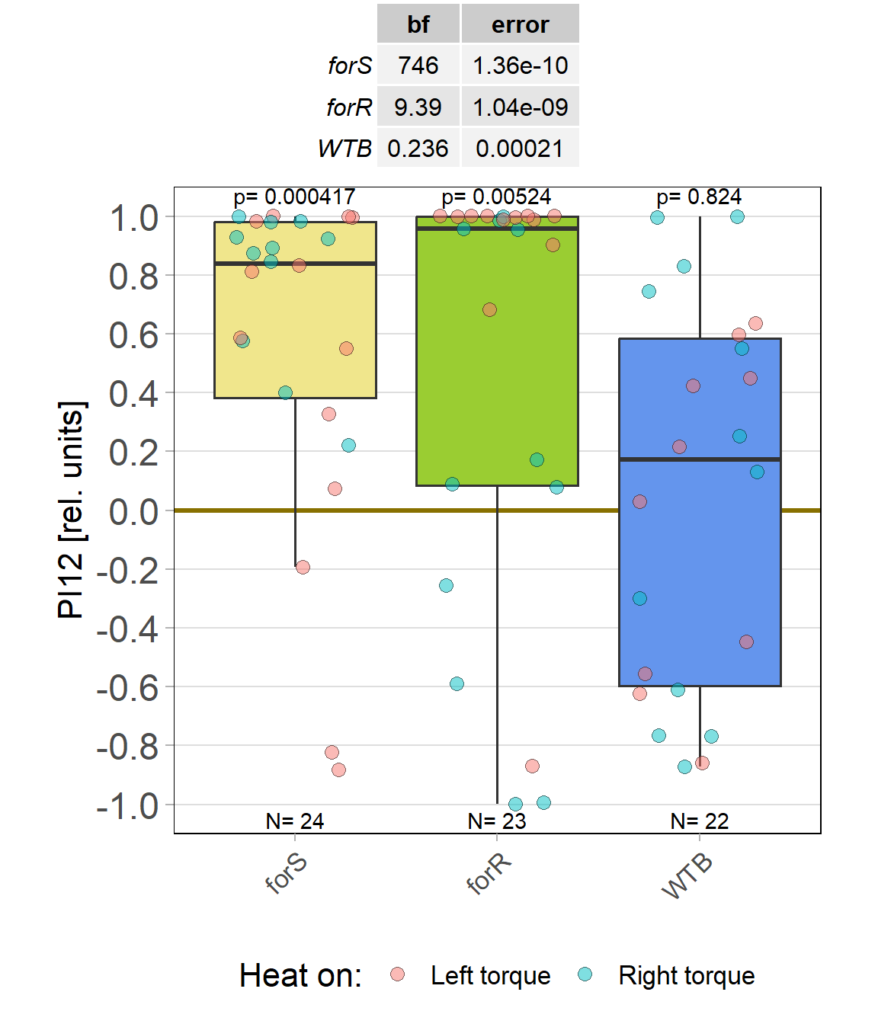
Self-learning, 8 minutes, elav-Gal4>UAS-for-RNAi
on Monday, July 8th, 2024 8:26 | by Radostina Lyutova
Category: Memory, Operant learning, operant self-learning, Rover/Sitter | No Comments
More data from the basement
on Monday, July 8th, 2024 8:15 | by Ellie
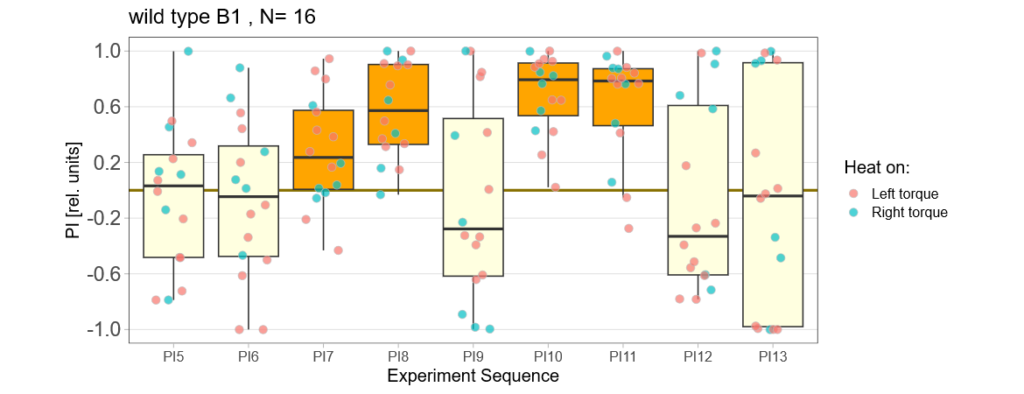
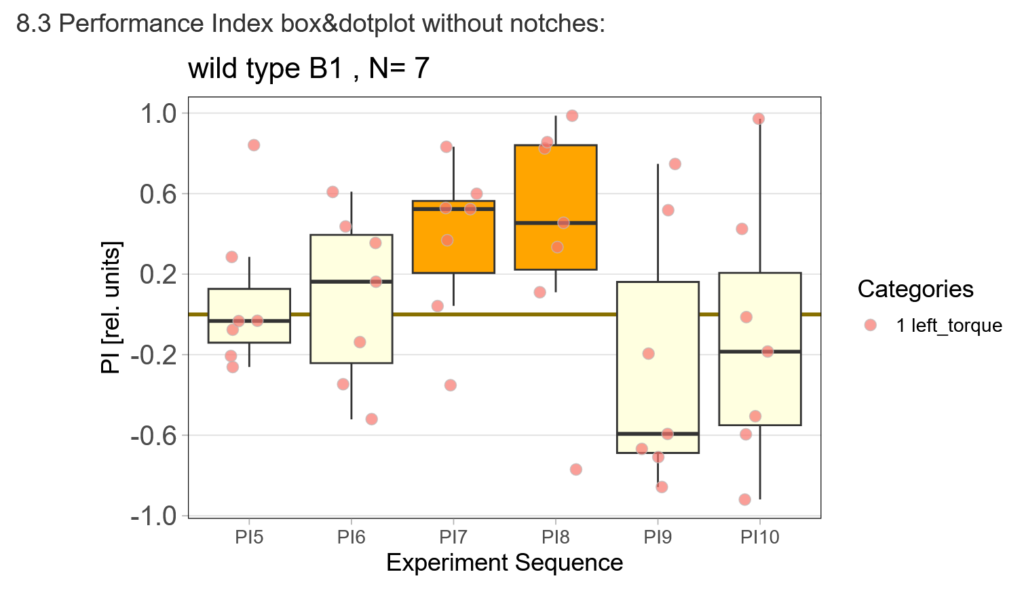
With the modified laser settings I was able to test 3 more wtb. Here are the results:

–> yaw torque
Testing the basement flight simulator
on Wednesday, June 19th, 2024 3:46 | by Ellie
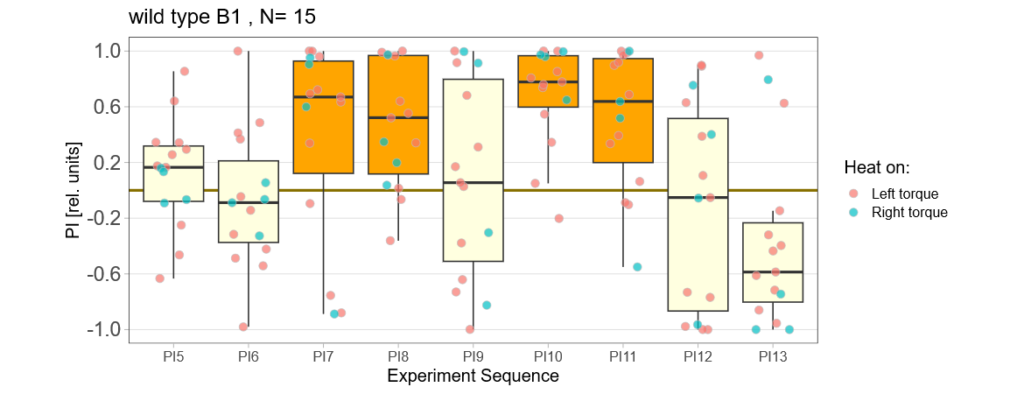
Before collecting the actual data I make sure that the flight simulator in the basement does its thing ;) Here are the test runs with wtb flies for different protocols:

—>> yaw torque

–>> switch mode

–>> habit formation
Rover vs. Sitter self learning after 4 minutes training
on Monday, October 16th, 2023 10:39 | by Radostina Lyutova
Category: flight, Habit formation, Memory, Operant learning, operant self-learning, Rover/Sitter | No Comments
Bachelor Blog / #7 offspring
on Monday, October 2nd, 2023 10:51 | by Ellie
Below you can find the data I collected from the offspring flies:
-> offspring from trained parents
-> offspring from untrained parents
(I left out data from flies that showed negative preference during two training periods in a row)
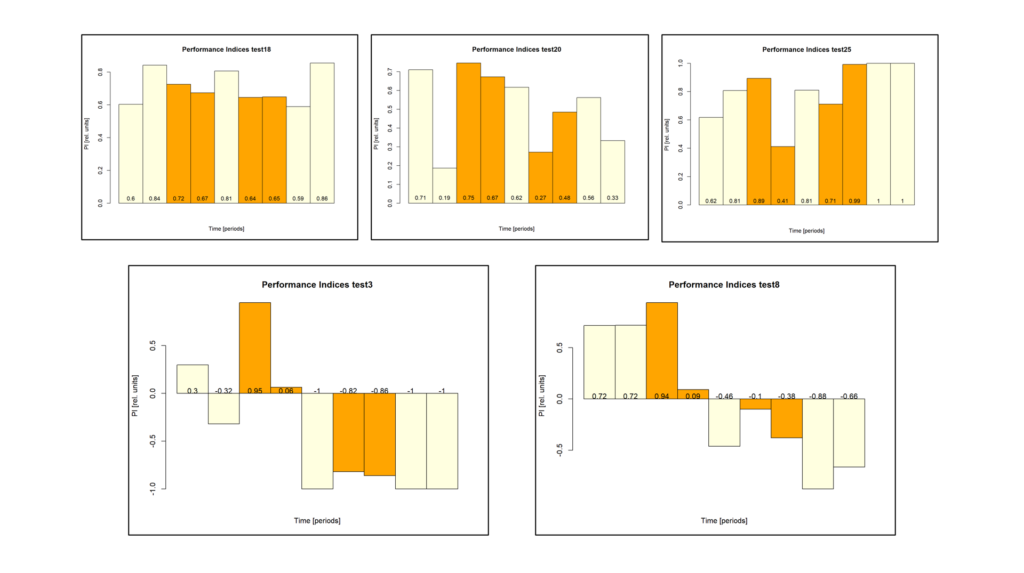
Bachelor Blog / #6 playing around
on Monday, September 18th, 2023 12:51 | by Ellie
Since the results I got after training the parental flies looked a bit odd on first sight I decided to take a closer view…
First I excluded some weird animals that either showed a larger preference for one side than avoidance or showed no avoidance two training periods in a row:
Next I compared the behavior of flies that showed avoidance but no learning with the behavior of the remaining flies:
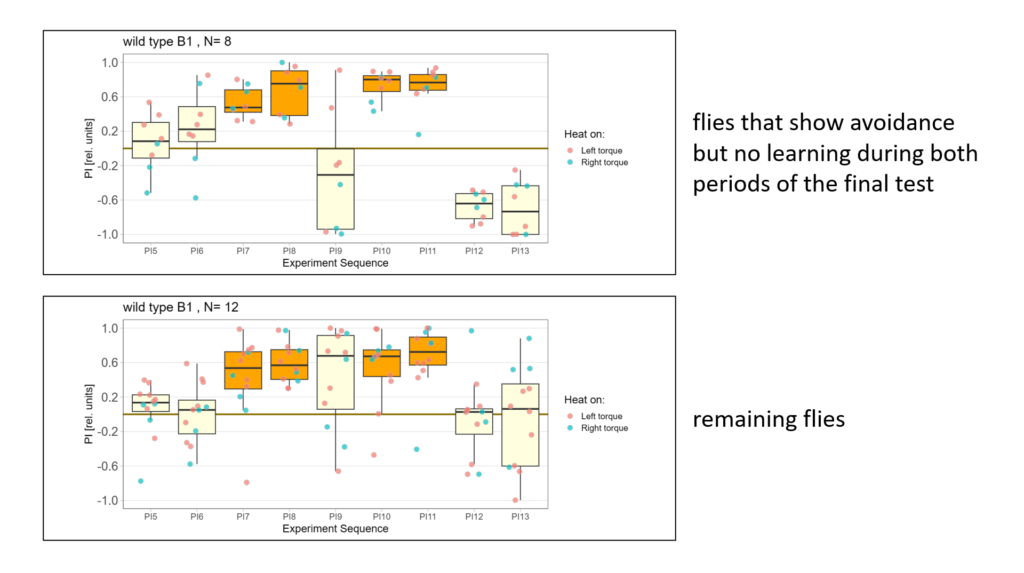
-> Avoidance is almost the same but note the first test period!
Lastly I split the data according to male and female flies. Here is what I got:
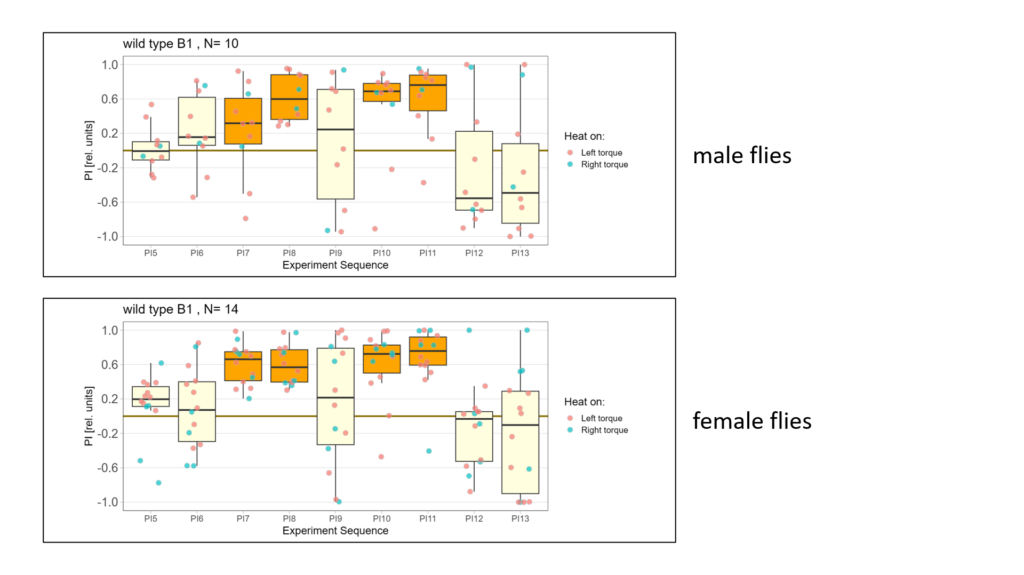
-> Looks a bit like there is negative learning in the male flies however I don´t have enough data to be sure…
As an overview here are all the flies (except for the excluded ones) together again:
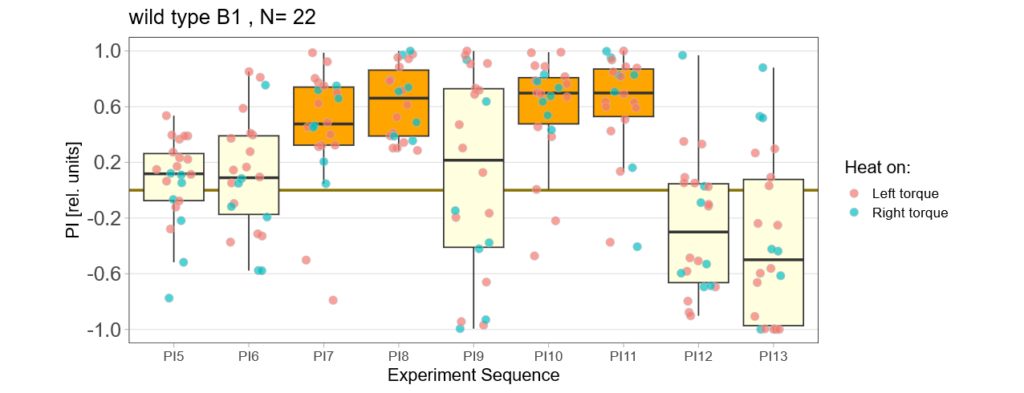
Bachelor Blog / #5 no learning :(
on Monday, September 11th, 2023 12:21 | by Ellie
Below you find the data from my experimental rounds A and B:
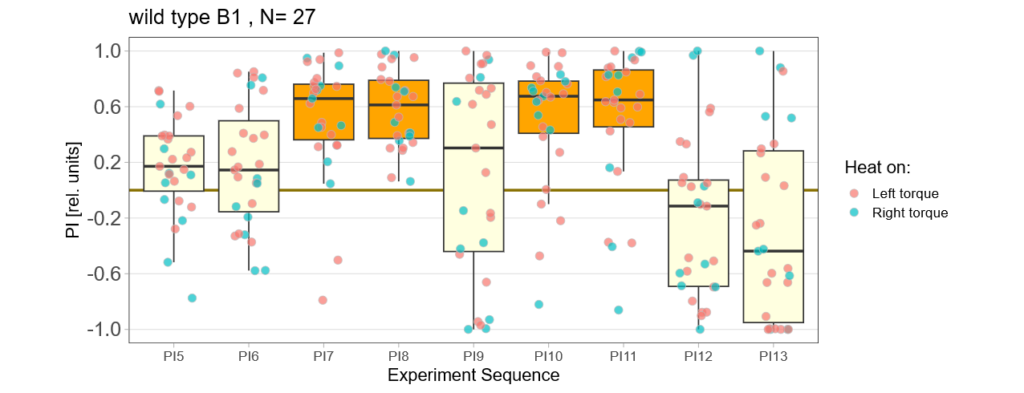
-> learning scores of the parental flies from experimental round A and B
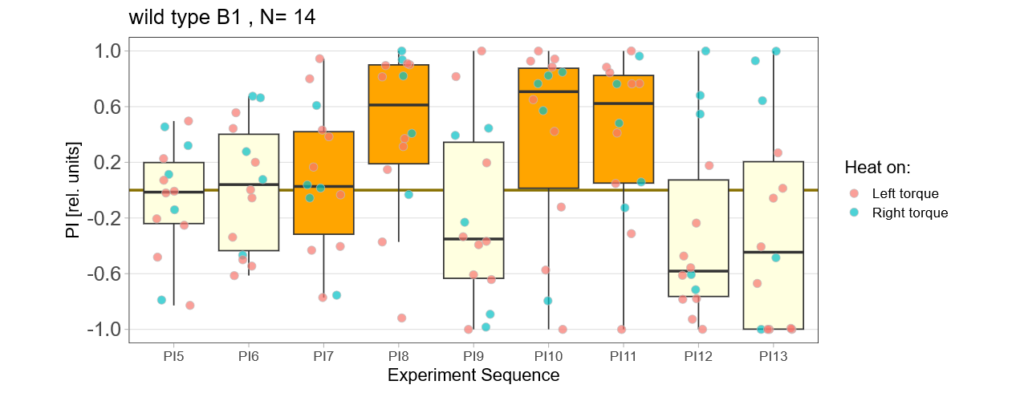
-> learning scores of the trained parent´s offspring only from experimental round A
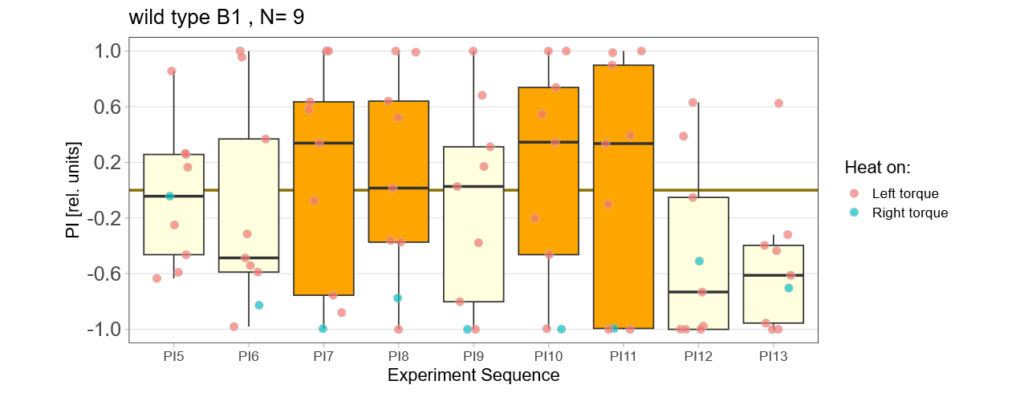
-> learning scores of the untrained parent´s offspring only from experimental round A
The results confuse me a lot and I am happy to discuss reasons :) However the offspring of the round-B will be ready for testing by the end of this week so there is still some data to collect…
Cloning via DNA Assembly
on Friday, September 1st, 2023 7:26 | by Isabel Stark
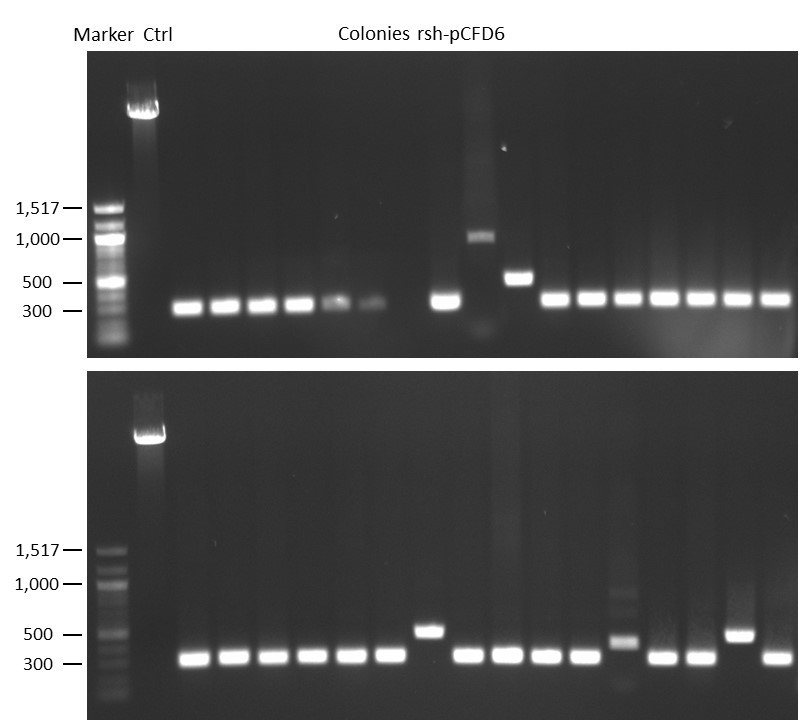
DNA Assembly in a 1:2 ratio of vector to insert with gRNAs of rsh and rut (Q5 and template concentration: 640 pg/µl) and 100ng of pCFD6 BbsI AP (using QuickCIP). Heat-shock (hs) transformation into E. coli (DH5α competent) with 10 µl Assembly Reaction and 100 µl cells.
For Crtl, pCFD6 BbsI AP was wrongly used.
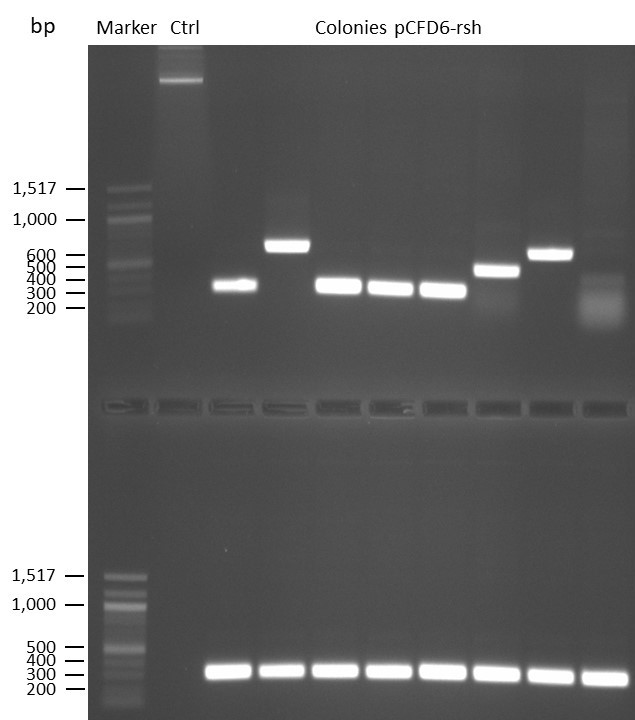
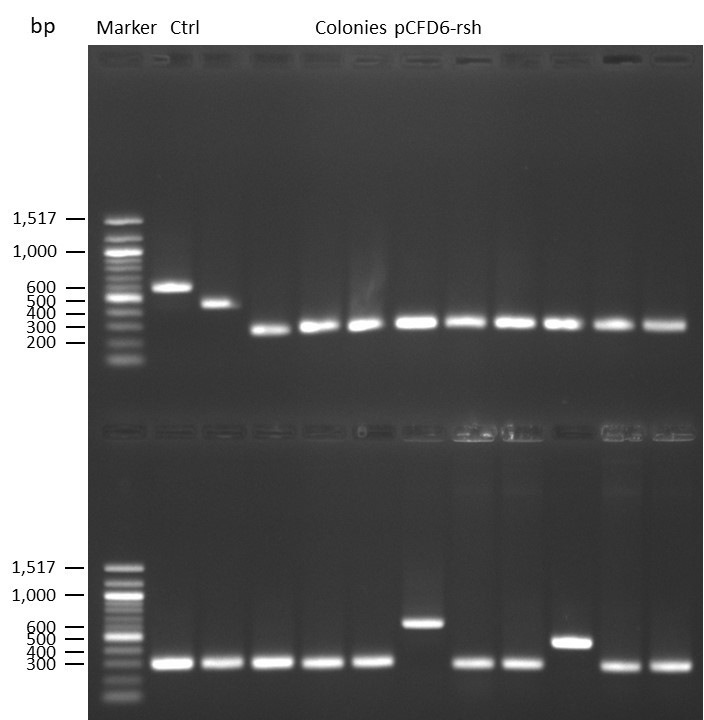
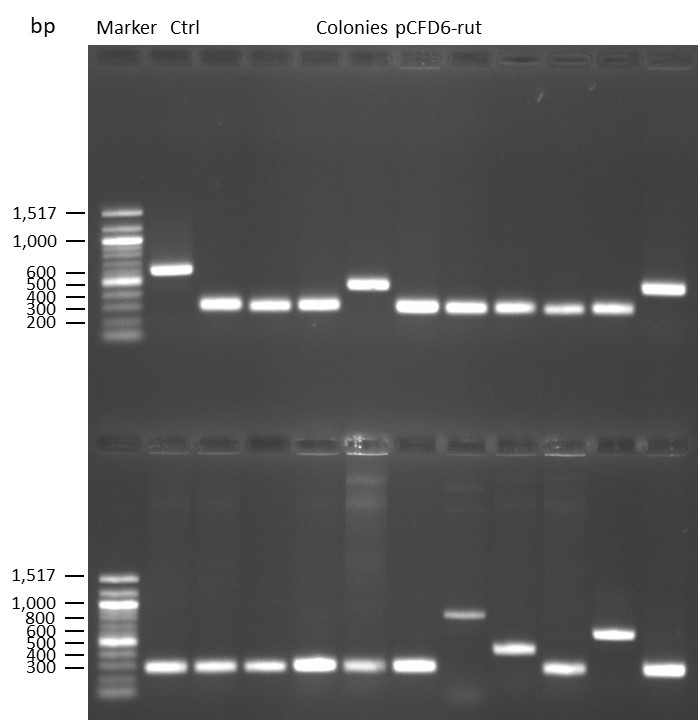
DNA Assembly in a 1:2 ratio of vector to insert with the gRNAs of rsh and rut (Q5 and template concentration: 640 pg/µl) and 100ng of pCFD6 BbsI AP (using FastAP and 2 extraction steps). Heat-shock (hs) transformation into E. coli (DH5α competent) with 10 µl Assembly Reaction and 100 µl cells.
For Crtl, pCFD6 BbsI AP was used in reaction.
Category: genetics, Memory, Operant learning, operant self-learning, Radish | No Comments
Bachelor Blog / #4 is there something?
on Monday, August 7th, 2023 2:13 | by Ellie
The offspring of my first experimental fly cohort finally hatched! Below you find a few first pre-tests I ran last week :)
First, here are the results of a quick test to see if the offspring shows a preference for the parentally trained side after the first training period:
After that I played around with the laser a little bit to find the learning threshhold. I set the laser on 2,6V but the results I got look a bit weird:
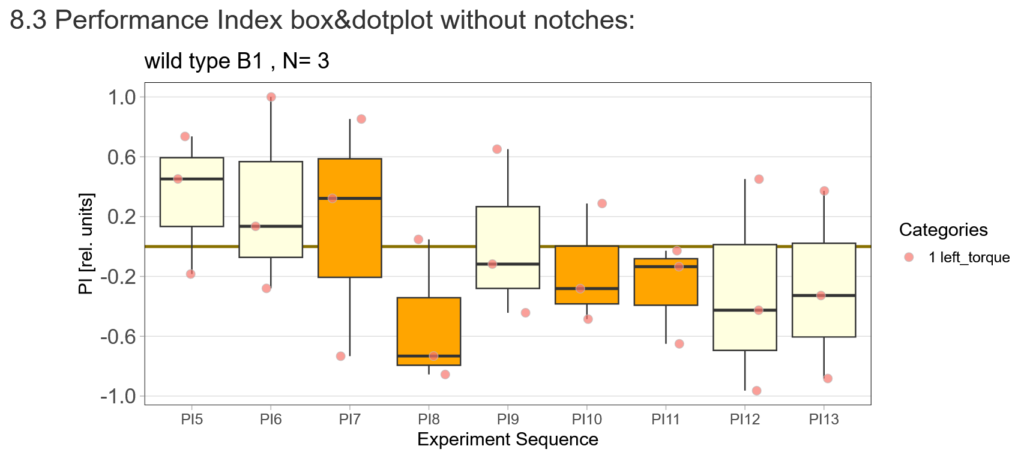
-> untrained wtb flies
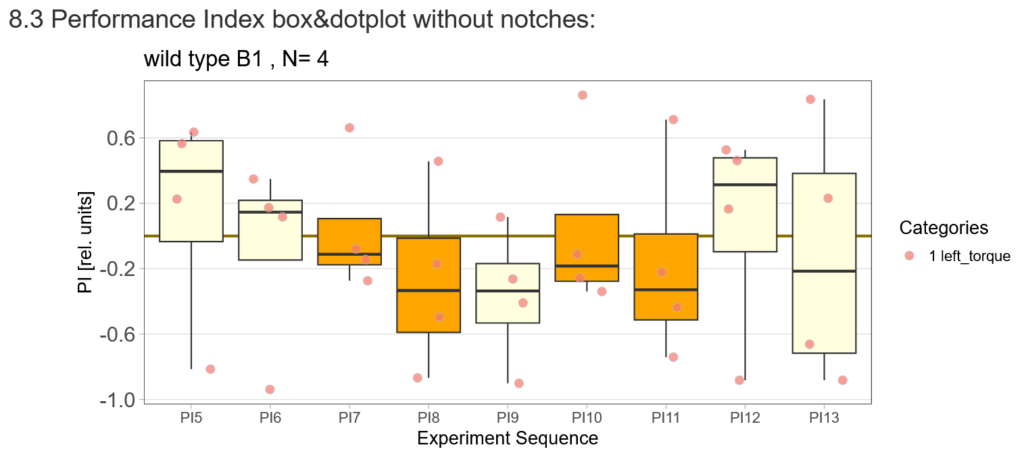
-> offspring of trained wtb flies
I´m optimistic however there is still a lot of work to do…
Success: rsh Stock has rsh1 Mutation
on Monday, August 7th, 2023 11:11 | by Isabel Stark
Via gDNA analysis and PCR was the specific area of the rsh gene extracted and amplified where the nucleotide substitution: C to T (Folkers et al., 2006) should be for the rsh1 mutation. The amplicon was Sanger sequenced which proved the nucleotide substitution.
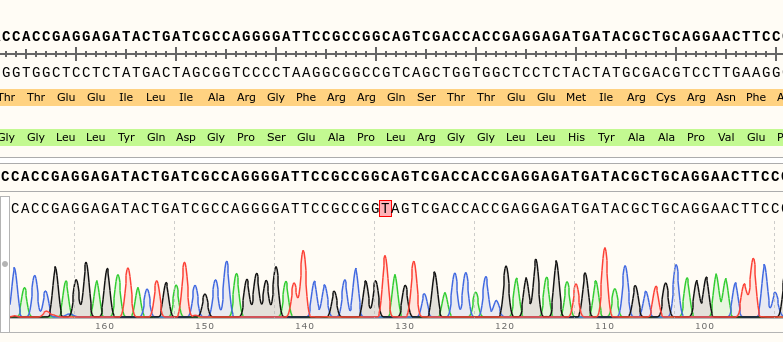
Category: genetics, Memory, Operant learning, operant self-learning, Radish, Uncategorized | No Comments