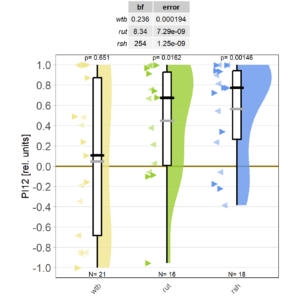
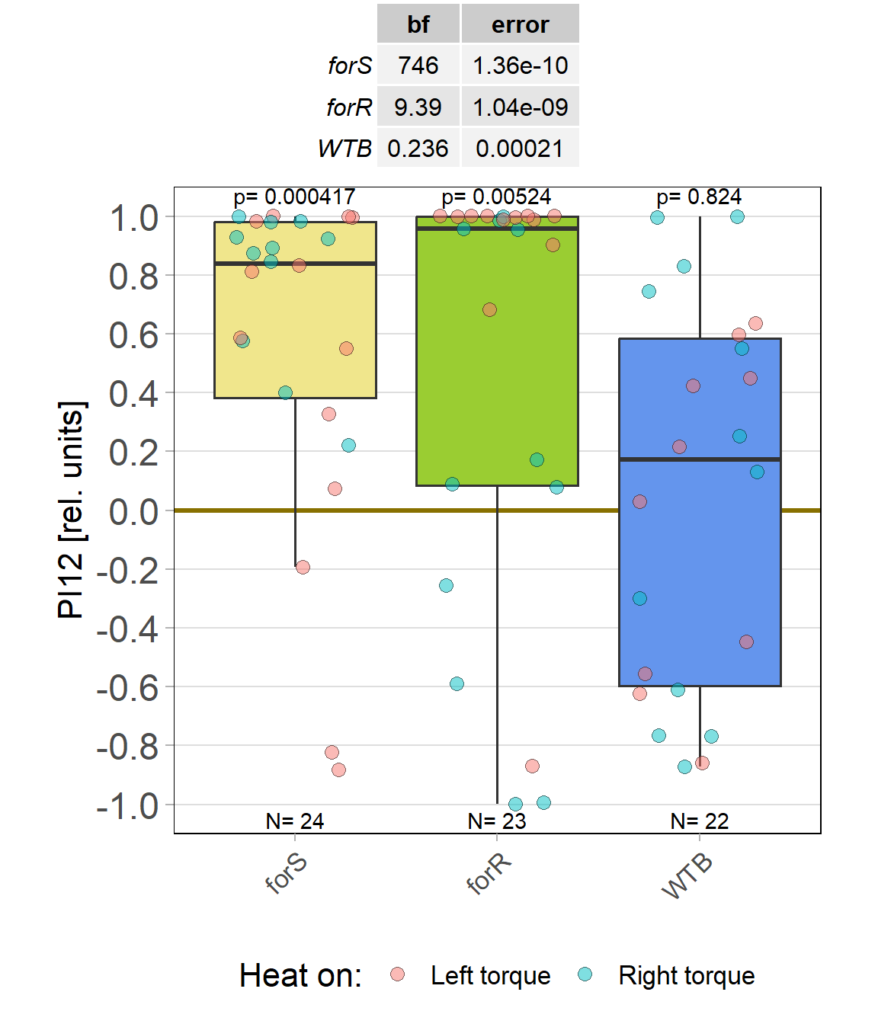
Starting it back up
on Monday, December 11th, 2023 11:34 | by Björn Brembs
Category: Operant learning, operant self-learning | No Comments
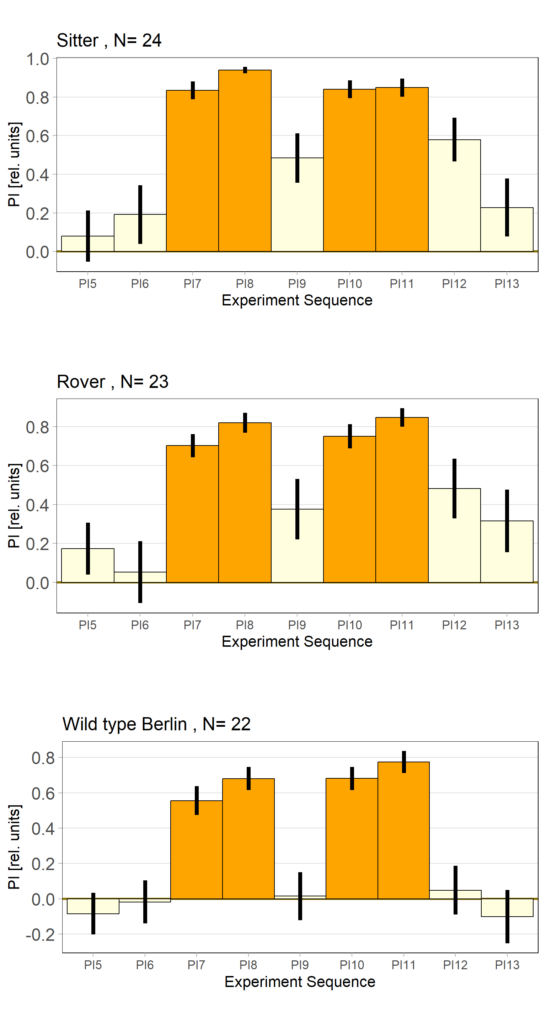
Rover vs. Sitter self learning after 4 minutes training
on Monday, October 16th, 2023 10:39 | by Radostina Lyutova
Category: flight, Habit formation, Memory, Operant learning, operant self-learning, Rover/Sitter | No Comments
Slowly collecting the mutants
on Friday, October 13th, 2023 6:13 | by Björn Brembs
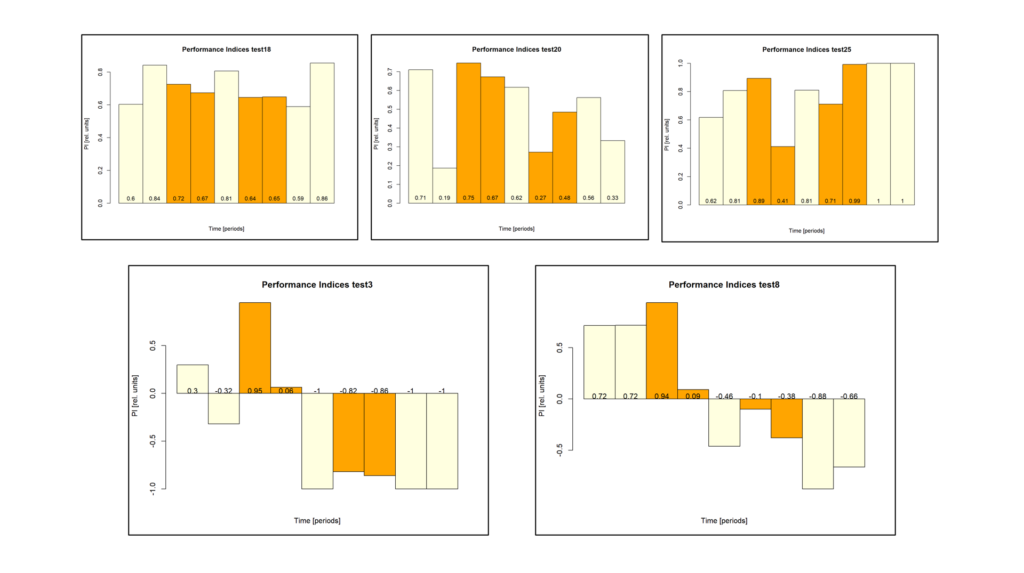
Finally, thanks to Marcella gluing to fly wheels instead of one, the mutant data are starting to roll in on the shortened self-learning experiment:
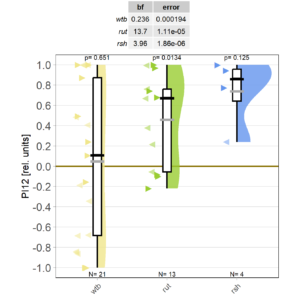
Category: Operant learning, operant self-learning | No Comments
First mutant data coming in
on Friday, October 6th, 2023 2:29 | by Björn Brembs
Short yaw torque learning, i.e., only one minute per period. Orange: training, yellow: test. WTB: wild type Berlin, rut: rutabaga learning mutants. With this short training of only 4 minutes, wild type flies show no torque preference in the after training, while at least the first few rutabaga flies show such a preference:
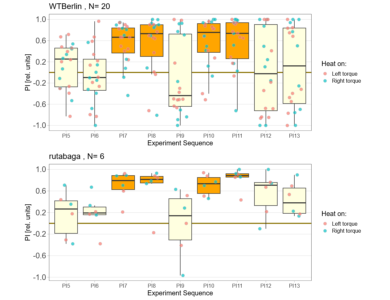
So far, I only could get one radish fly to make it through the experiment, so I cannot display it here.
Category: Operant learning, operant self-learning | No Comments
Bachelor Blog / #7 offspring
on Monday, October 2nd, 2023 10:51 | by Ellie
Below you can find the data I collected from the offspring flies:
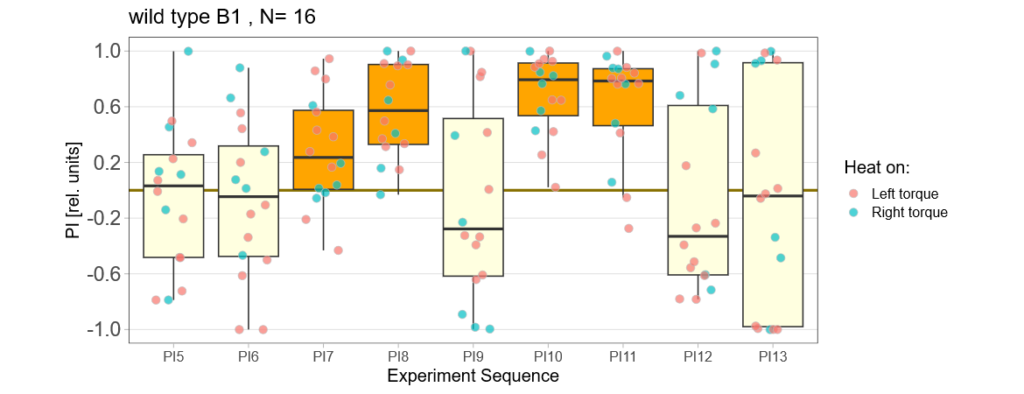
-> offspring from trained parents
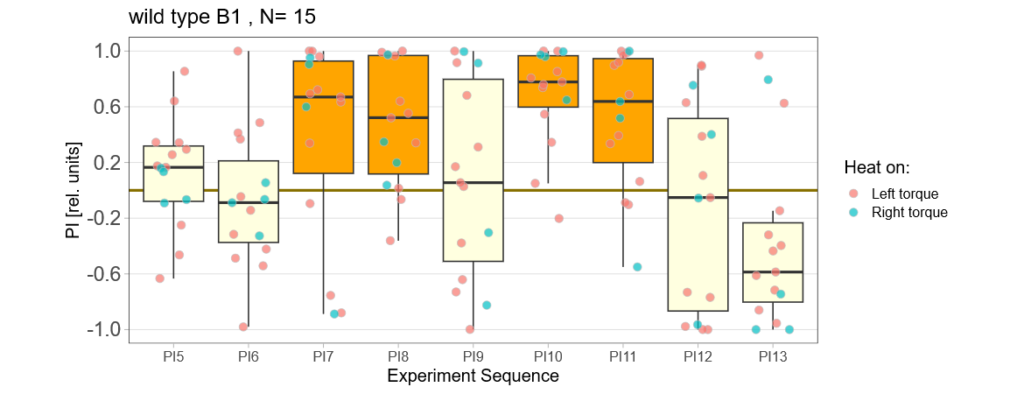
-> offspring from untrained parents
(I left out data from flies that showed negative preference during two training periods in a row)
Four minutes not enough
on Friday, September 29th, 2023 4:20 | by Björn Brembs
Eight minutes of yaw torque training work just fine for both wild type and mutant flies:
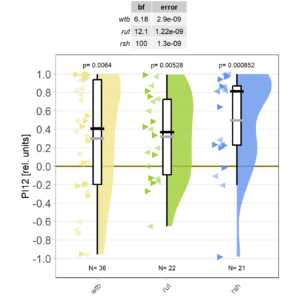
Reducing the training to four minutes is not enough for wild type flies:
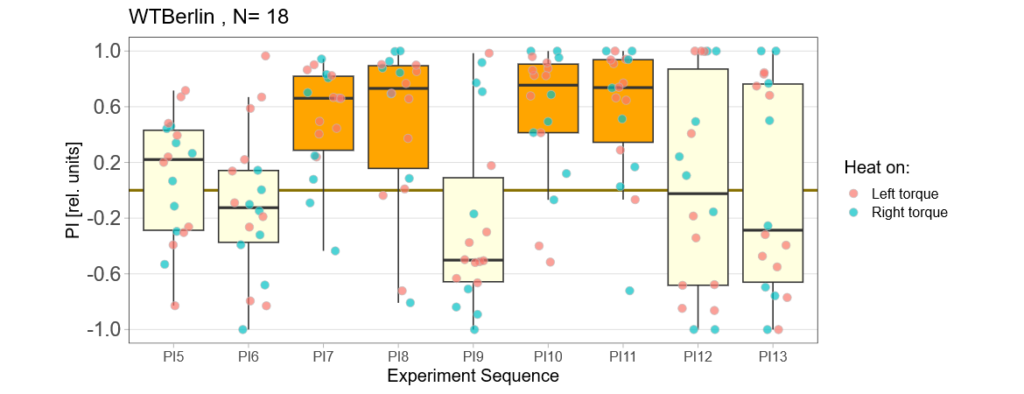
Now it will be exciting to see if the mutants still do what they did many years ago: learn better than wild type.
Category: Operant learning, operant self-learning | No Comments
Results for MB+DANs and MB054B
on Monday, September 25th, 2023 11:07 | by Maja Achatz
The first experiment tested the activation of KCs with simultaneous ablation of pPAM which showed the following results:
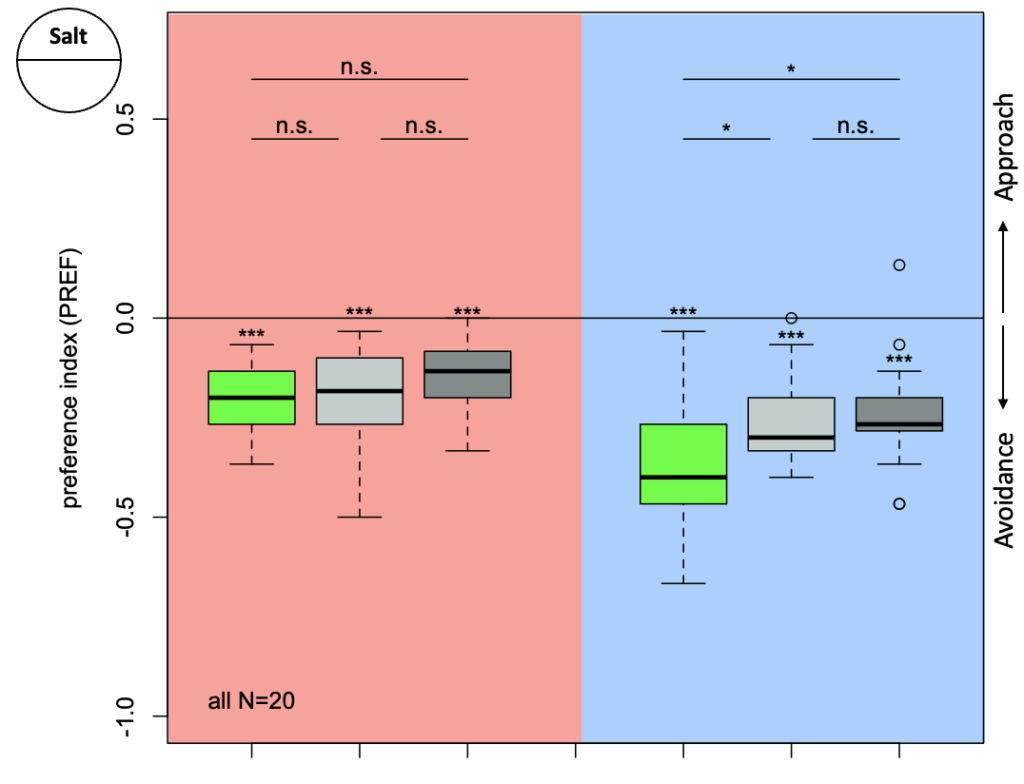
Green: UAS-ChR2-XXL; LexAop-rpr x 58E02-LexA; H24-Gal4
Lightgrey: UAS-ChR2-XXL; LexAop-rpr x H24-Gal4
Darkgrey: UAS-ChR2-XXL x 58E02-LexA; H24-Gal4
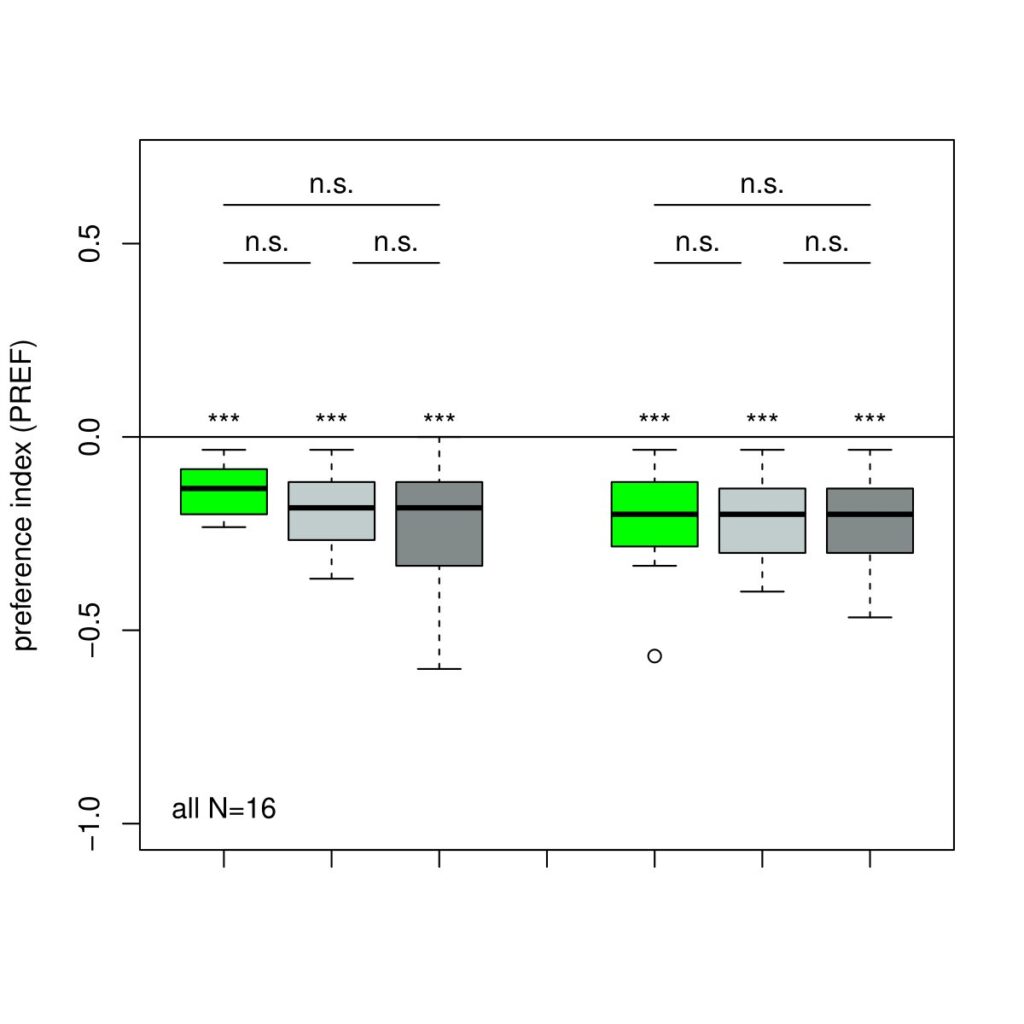
The next experiment was conducted with the split-Gal4 line MB054B (DAN-f1/DAN-g1) and showed the following:
Category: Kenyon cells, Larve, Mushroom Body, Optogenetics | No Comments
Bachelor Blog / #6 playing around
on Monday, September 18th, 2023 12:51 | by Ellie
Since the results I got after training the parental flies looked a bit odd on first sight I decided to take a closer view…
First I excluded some weird animals that either showed a larger preference for one side than avoidance or showed no avoidance two training periods in a row:
Next I compared the behavior of flies that showed avoidance but no learning with the behavior of the remaining flies:
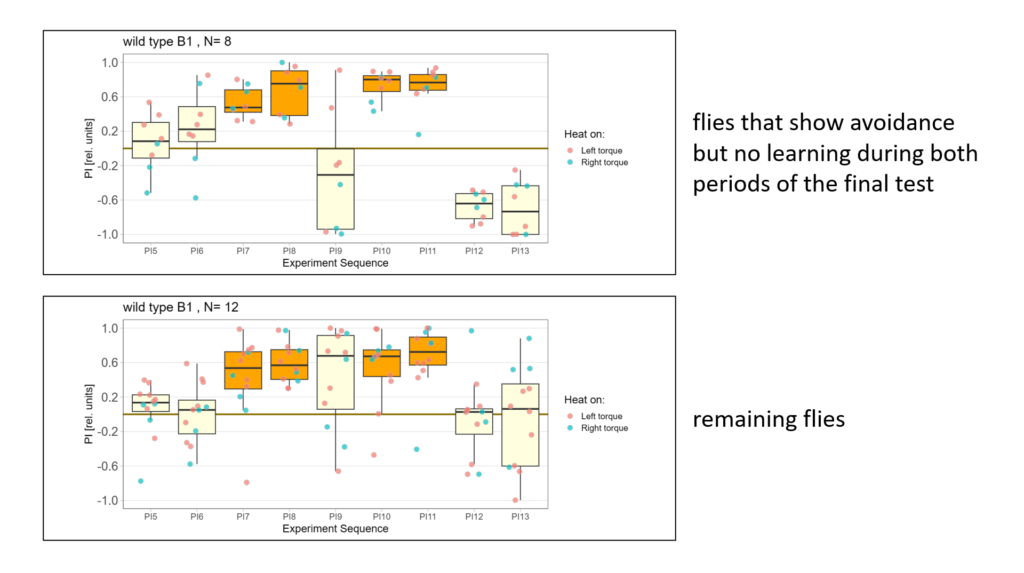
-> Avoidance is almost the same but note the first test period!
Lastly I split the data according to male and female flies. Here is what I got:
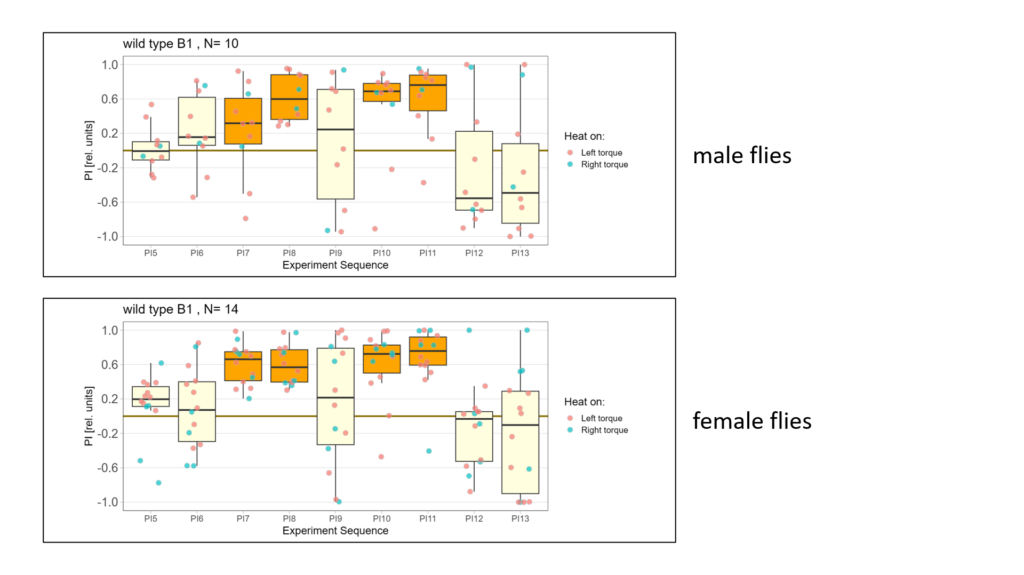
-> Looks a bit like there is negative learning in the male flies however I don´t have enough data to be sure…
As an overview here are all the flies (except for the excluded ones) together again:
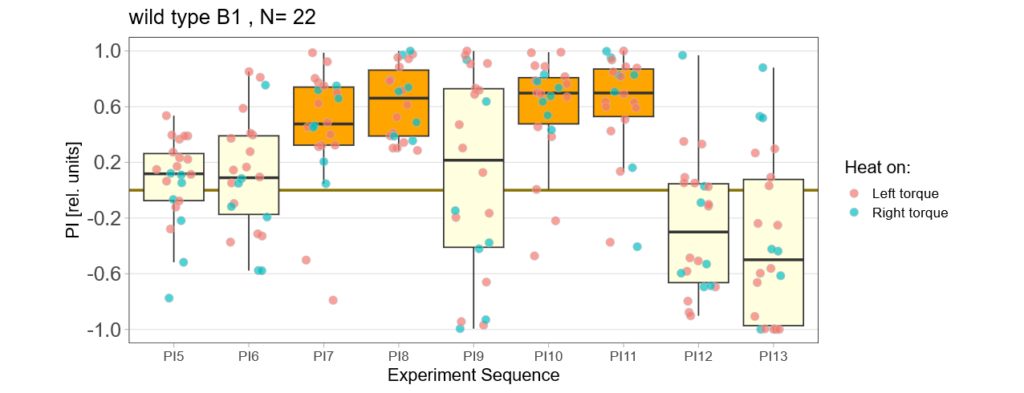
Bachelor Blog / #5 no learning :(
on Monday, September 11th, 2023 12:21 | by Ellie
Below you find the data from my experimental rounds A and B:
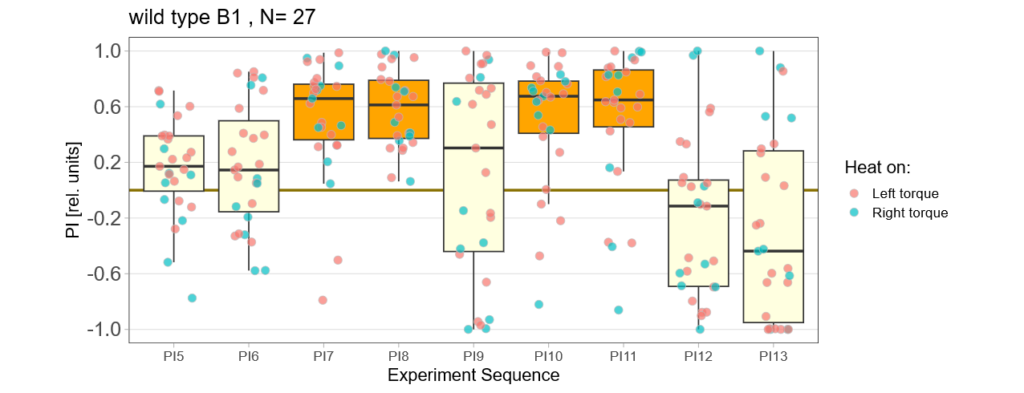
-> learning scores of the parental flies from experimental round A and B
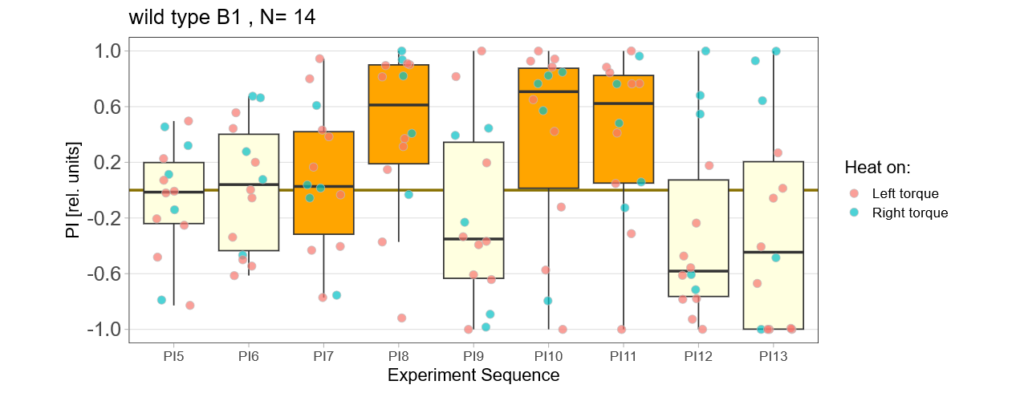
-> learning scores of the trained parent´s offspring only from experimental round A
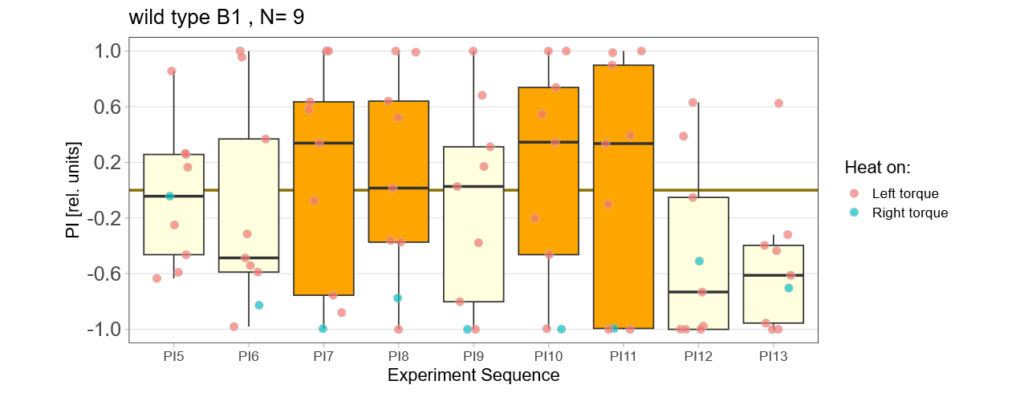
-> learning scores of the untrained parent´s offspring only from experimental round A
The results confuse me a lot and I am happy to discuss reasons :) However the offspring of the round-B will be ready for testing by the end of this week so there is still some data to collect…
Verification TH-C-AD;TH-D-DBD
on Monday, September 11th, 2023 9:52 | by Luisa Guyton
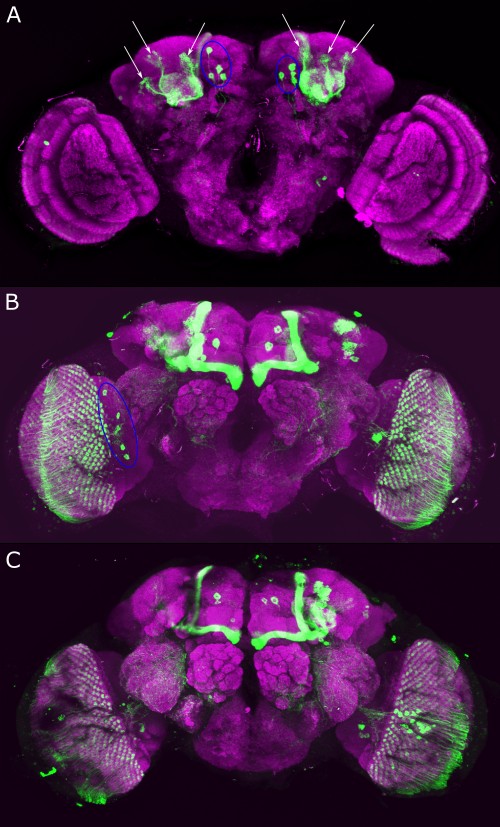
Category: Anatomy, DAN, genetics, Kenyon cells, Optogenetics | No Comments