Verification TH-C-AD;TH-D-DBD
on Monday, September 11th, 2023 9:52 | by Luisa Guyton
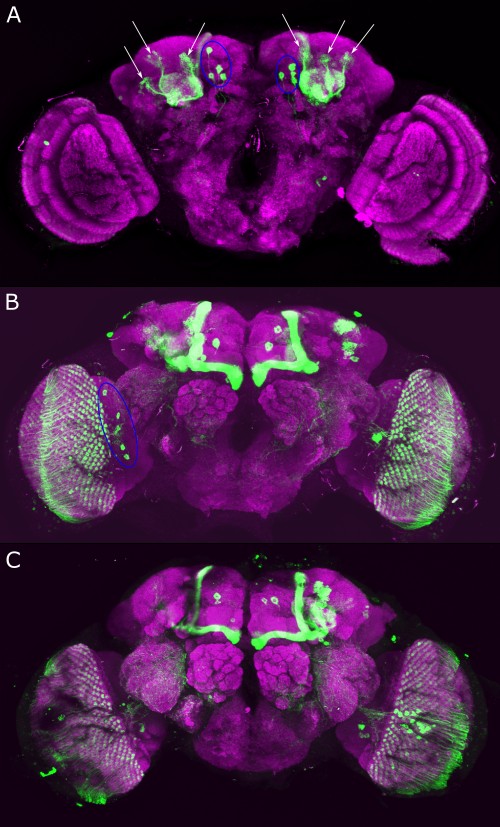
Category: Anatomy, DAN, genetics, Kenyon cells, Optogenetics | No Comments
Cloning via DNA Assembly
on Friday, September 1st, 2023 7:26 | by Isabel Stark
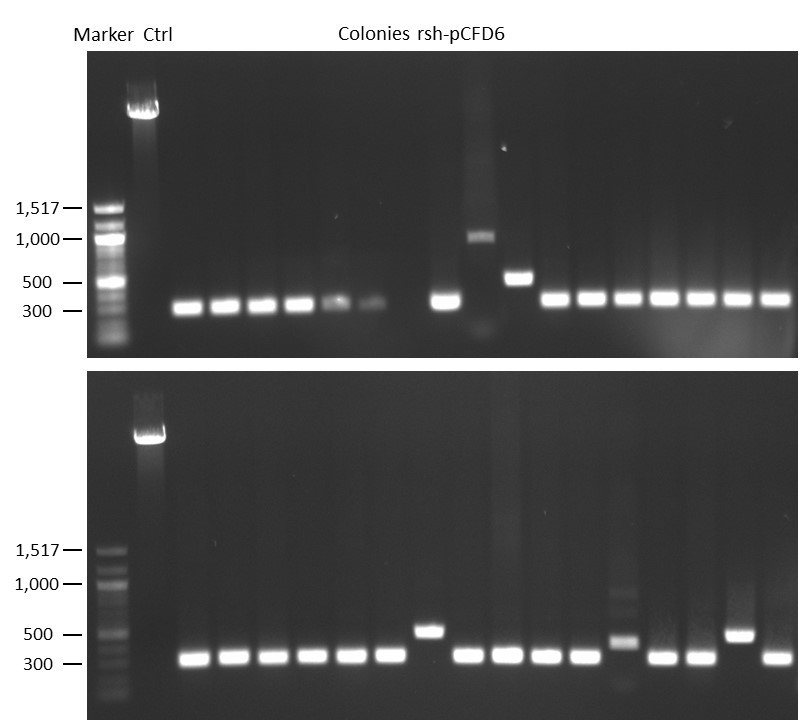
DNA Assembly in a 1:2 ratio of vector to insert with gRNAs of rsh and rut (Q5 and template concentration: 640 pg/µl) and 100ng of pCFD6 BbsI AP (using QuickCIP). Heat-shock (hs) transformation into E. coli (DH5α competent) with 10 µl Assembly Reaction and 100 µl cells.
For Crtl, pCFD6 BbsI AP was wrongly used.
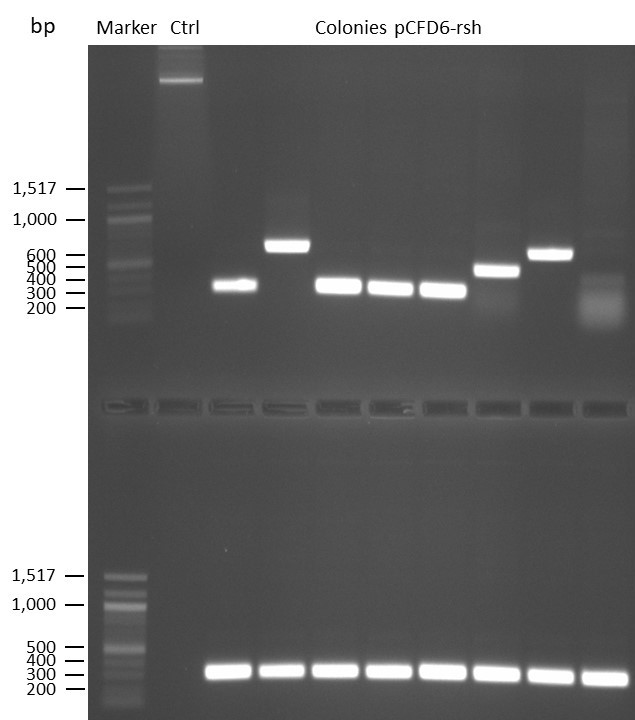
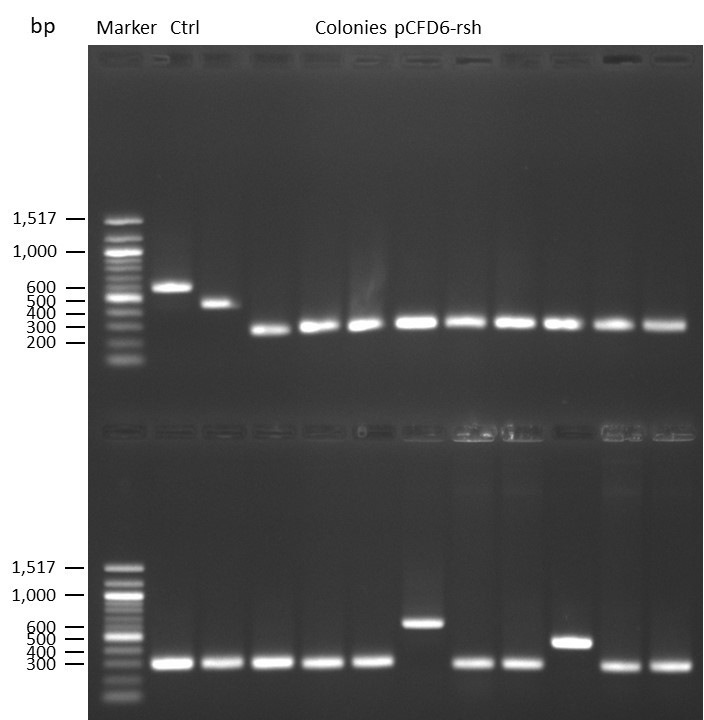
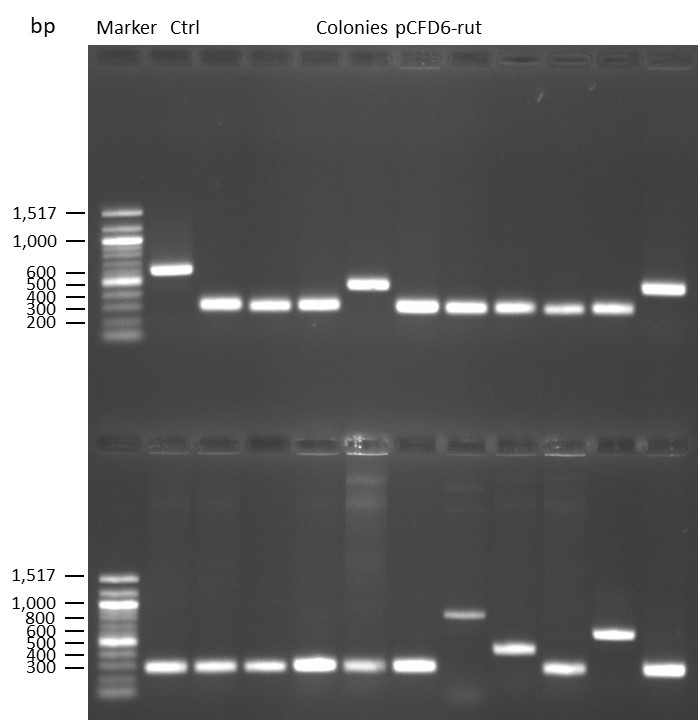
DNA Assembly in a 1:2 ratio of vector to insert with the gRNAs of rsh and rut (Q5 and template concentration: 640 pg/µl) and 100ng of pCFD6 BbsI AP (using FastAP and 2 extraction steps). Heat-shock (hs) transformation into E. coli (DH5α competent) with 10 µl Assembly Reaction and 100 µl cells.
For Crtl, pCFD6 BbsI AP was used in reaction.
Category: genetics, Memory, Operant learning, operant self-learning, Radish | No Comments
Line verification SS56699
on Tuesday, August 29th, 2023 7:50 | by Luisa Guyton
After dissecting brains from the GAL4 driver line SS56699 with GFP staining and finding no fluorescence, I dissected them again and the immunohistochemical staining showed fluorescence. To check that the correct neurons were stained, I compared the images with the image in the paper by Hulse et al (2021; https://doi.org/10.7554/eLife.66039) . The three PPL1 dopaminergic dorsal fan-shaped body tangential neurons per hemisphere are stained, but there are some additional unidentified neurons (presumably PPM1 neruons) visible.
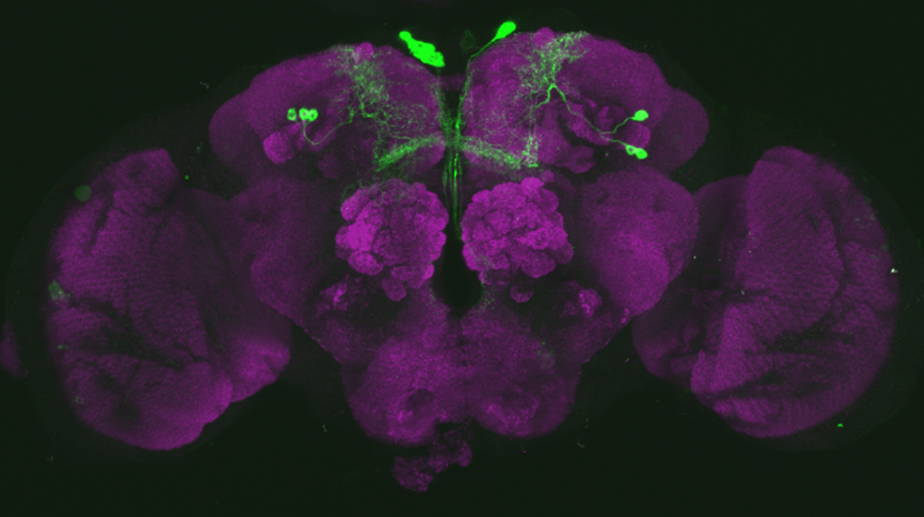
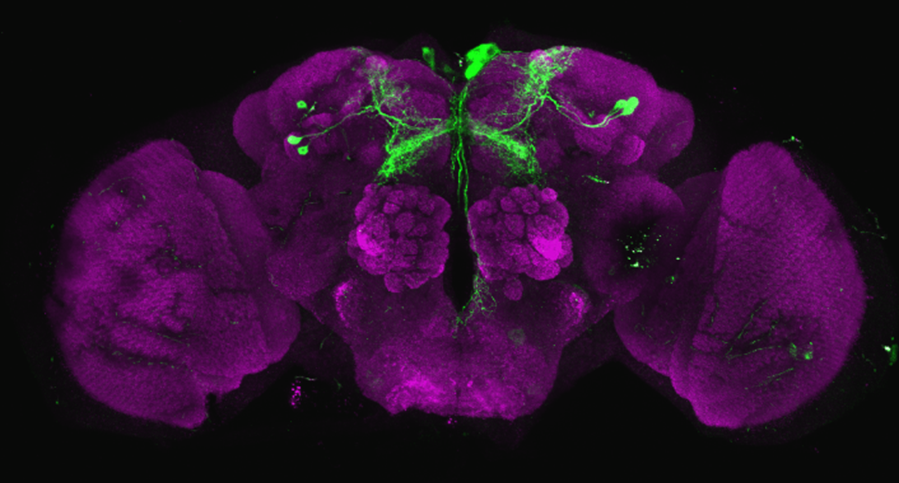
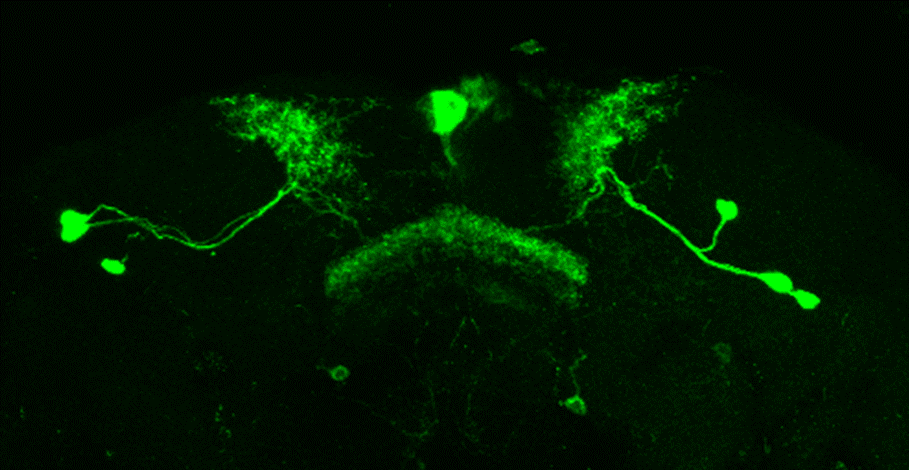
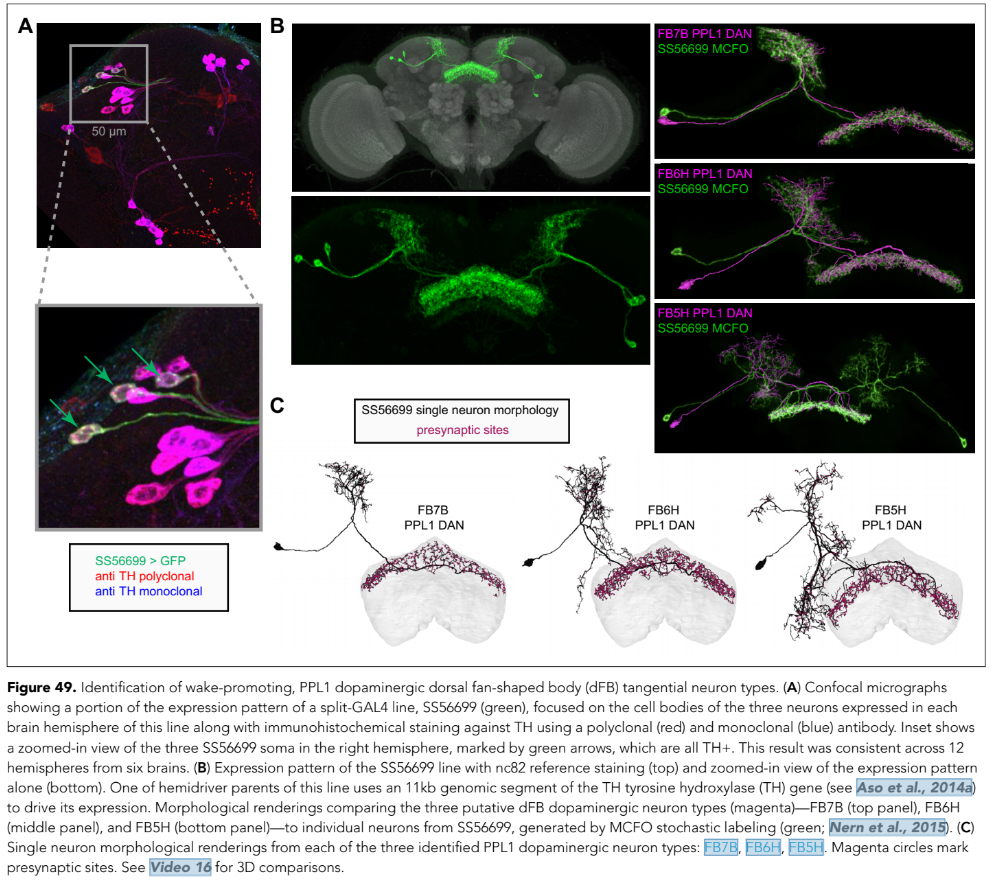
Hulse, B. K., Haberkern, H., Franconville, R., Turner-Evans, D., Takemura, S.-y., Wolff, T., Noorman, M., Dreher, M., Dan, C., Parekh, R., Hermundstad, A. M., Rubin, G. M., & Jayaraman, V. (2021). A connectome of the Drosophila central complex reveals network motifs suitable for flexible navigation and context-dependent action selection. eLife, 10, e66039. https://doi.org/10.7554/eLife.66039
Category: Anatomy, genetics | No Comments
Success: rsh Stock has rsh1 Mutation
on Monday, August 7th, 2023 11:11 | by Isabel Stark
Via gDNA analysis and PCR was the specific area of the rsh gene extracted and amplified where the nucleotide substitution: C to T (Folkers et al., 2006) should be for the rsh1 mutation. The amplicon was Sanger sequenced which proved the nucleotide substitution.
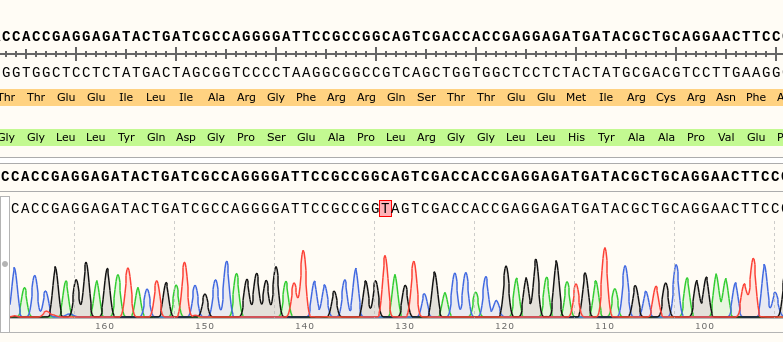
Category: genetics, Memory, Operant learning, operant self-learning, Radish, Uncategorized | No Comments
Creating gRNAs via PCR
on Friday, July 7th, 2023 6:38 | by Isabel Stark
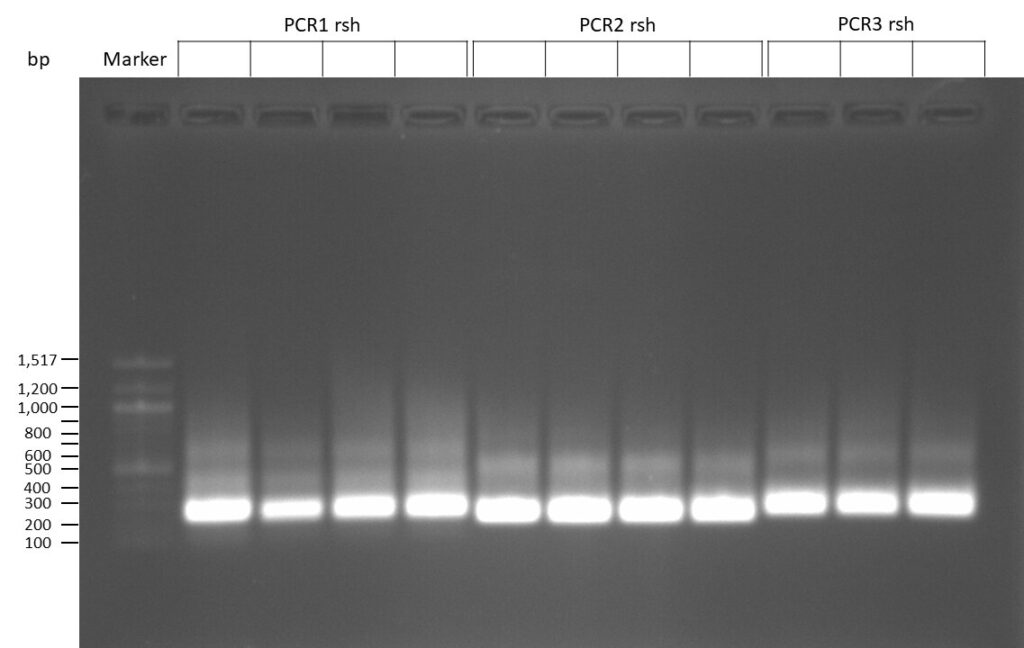
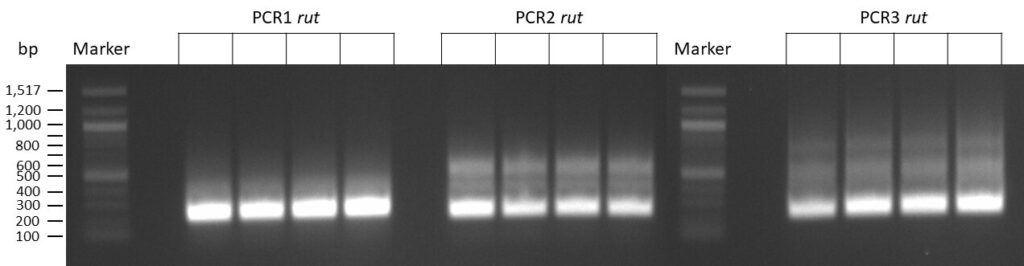
The template pCFD6 was used with a concentration of 640 pg/µl.
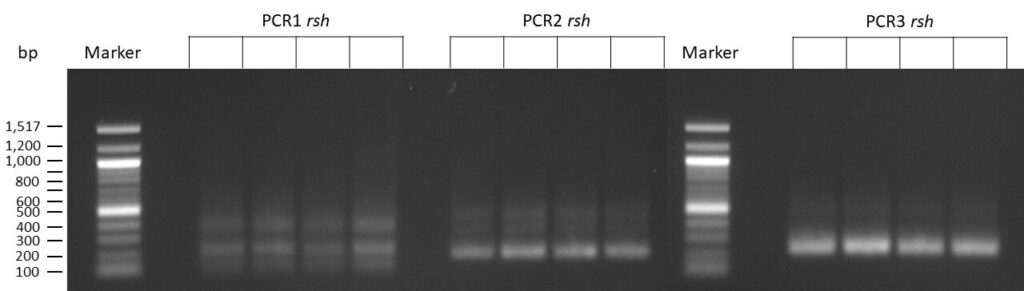
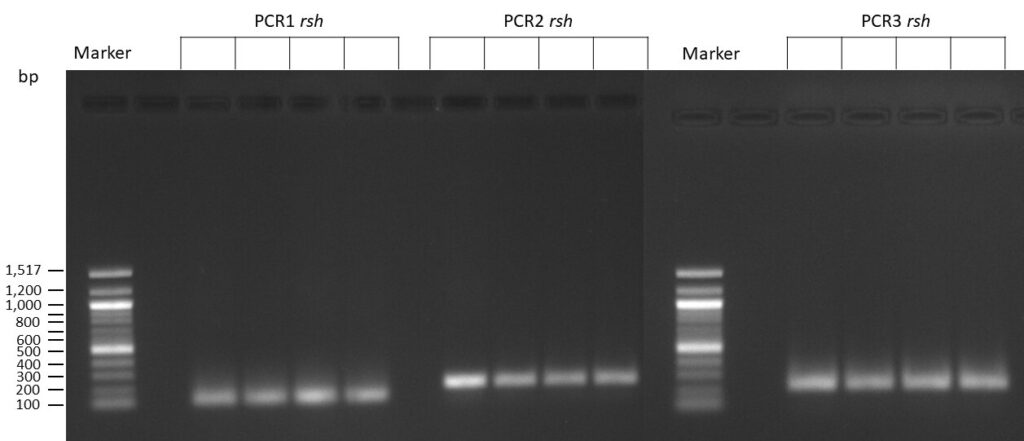
The template pCFD6 was used with a concentration of 64 pg/µl (1:10 dilution).
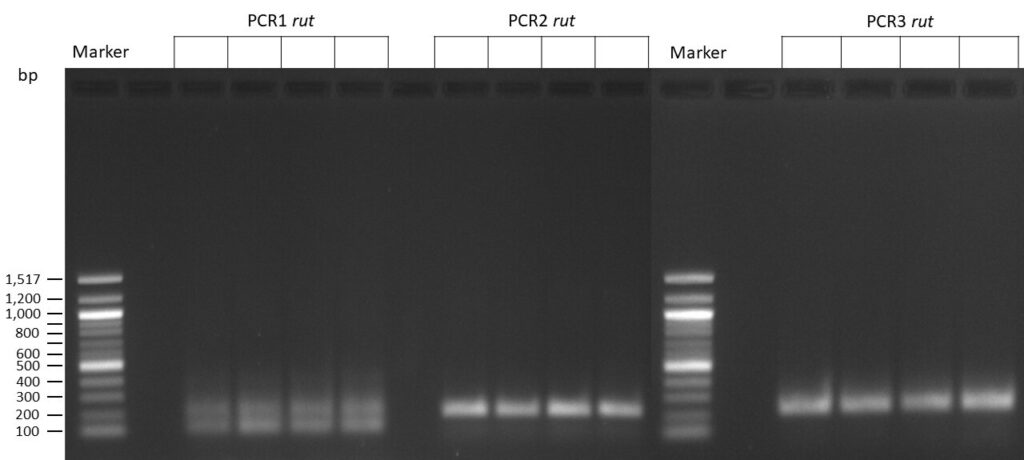
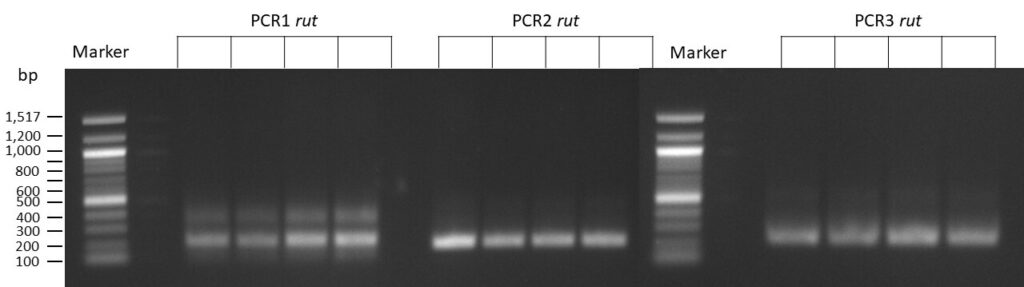
The template pCFD6 was used with a concentration of 64 pg/µl (1:10 dilution).
50µl of 5xQ5 High GC Enhancer was added to the PCR mix.
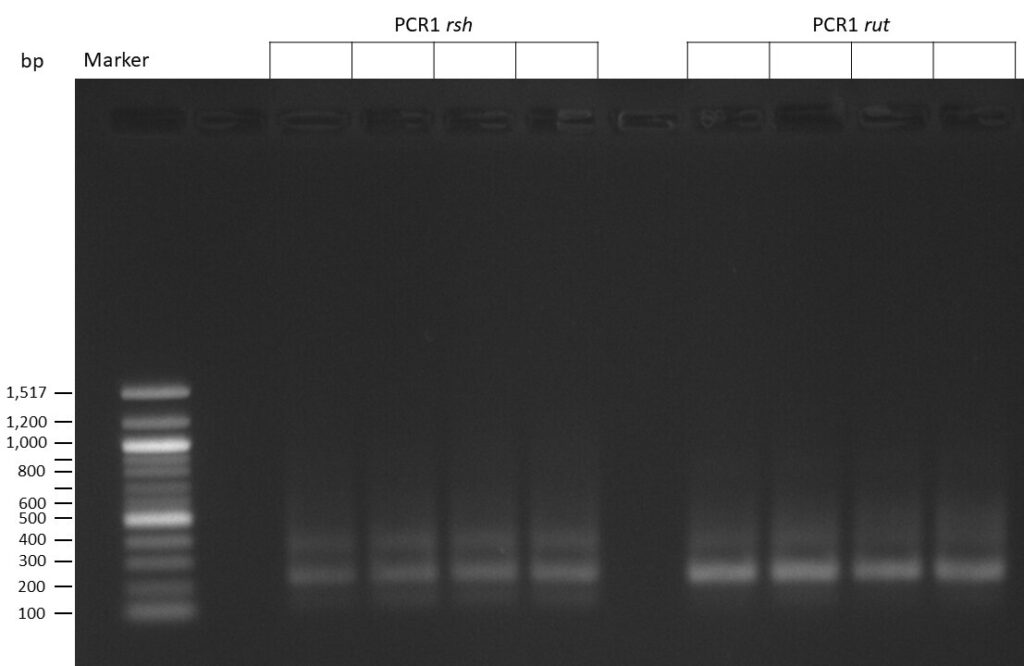
The template pCFD6 was used with a concentration of 128 pg/µl (1:5 dilution).
50µl of 5xQ5 High GC Enhancer was added to the PCR mix.
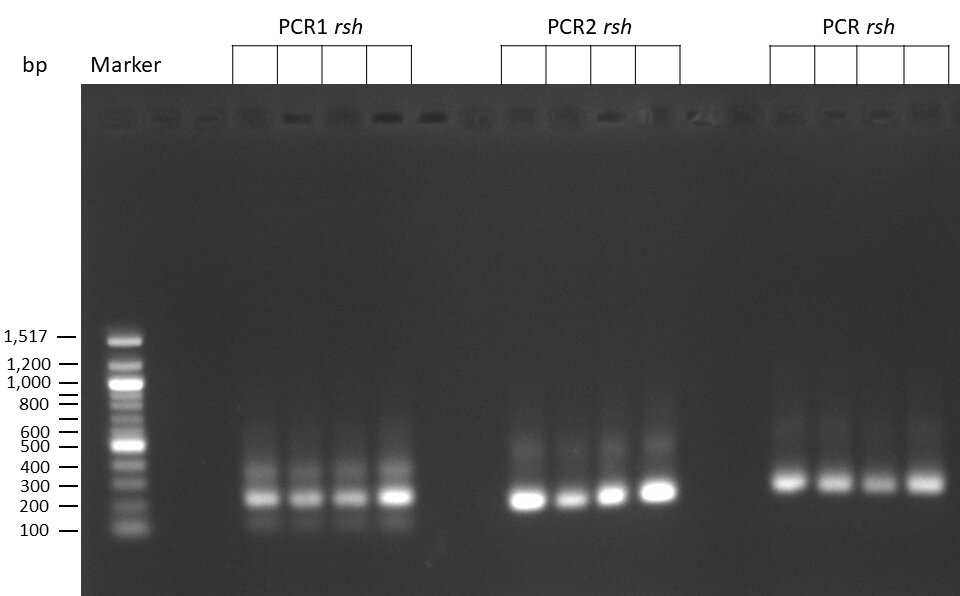
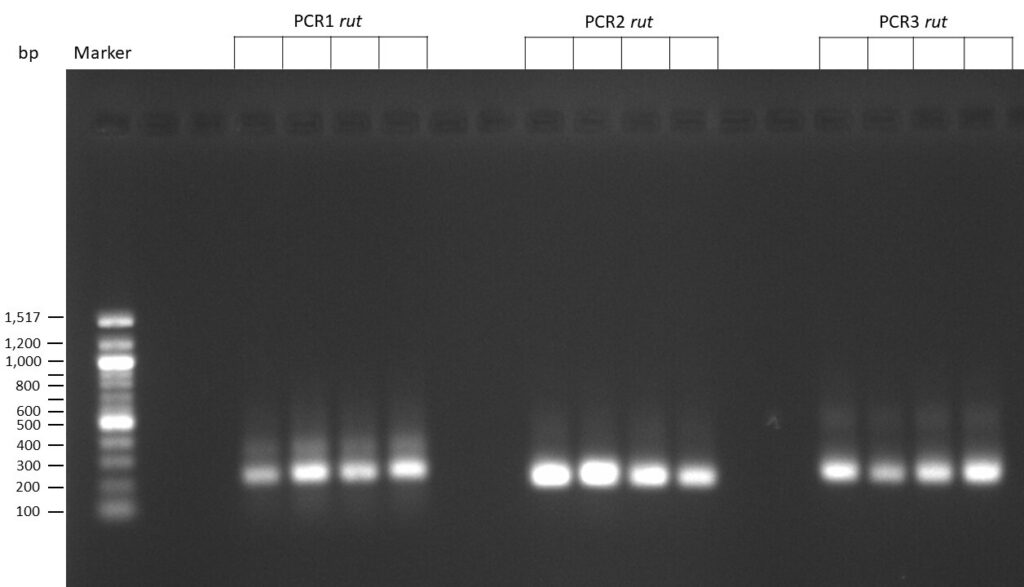
The template pCFD6 was used with a concentration of 640 pg/µl.
The Phusion DNA Polymerase was used instead of the Q5 High-Fidelity DNA Polymerase.
Category: genetics, Memory, Operant learning, operant self-learning, Radish, Uncategorized | No Comments
Gene switch finally worked
on Monday, September 28th, 2020 12:21 | by Andreas Ehweiner
I retried the gene-switch method with a new solution of RU486. Since it did not worked before I just tried one line. The flies were left for two days on instant food mixed with a sugar solution of RU486. This time I saw normal GFP expression. So I will test all 4 line in parallel after the break.
Category: Anatomy, crosses | No Comments
Testing of possible Gal4-lines
on Monday, September 28th, 2020 12:11 | by Andreas Ehweiner
I finished my cross for the colocalisation of FoxP with the 6 Gal4-lines i ordered.
Two lines show a nice colocalisation and will be tested.

One would show some overlap, but a completly diffrent expression pattern than it should have. So i will try this line again from the stock to exclude a mixup of the line.
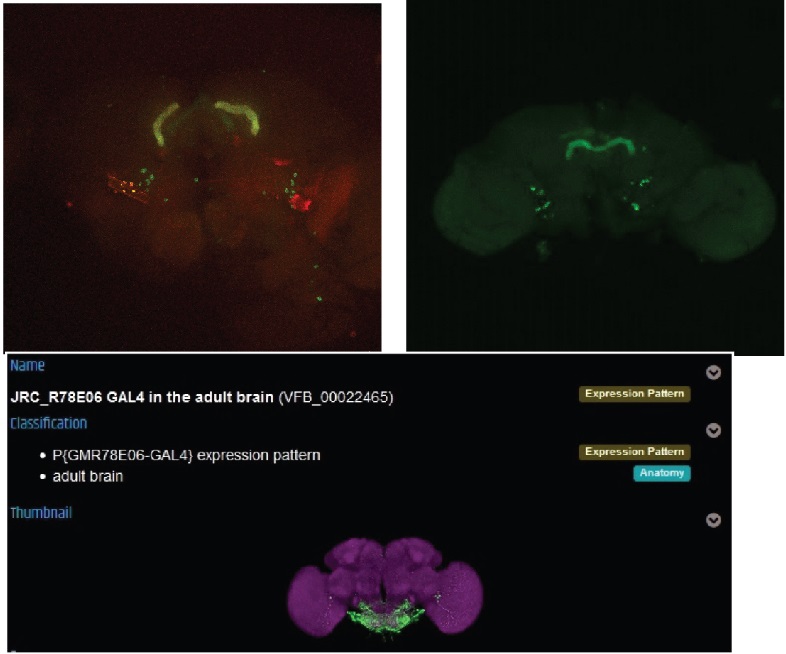
The next one dont seem to have any overlap, but also the pattern looks not like it should.
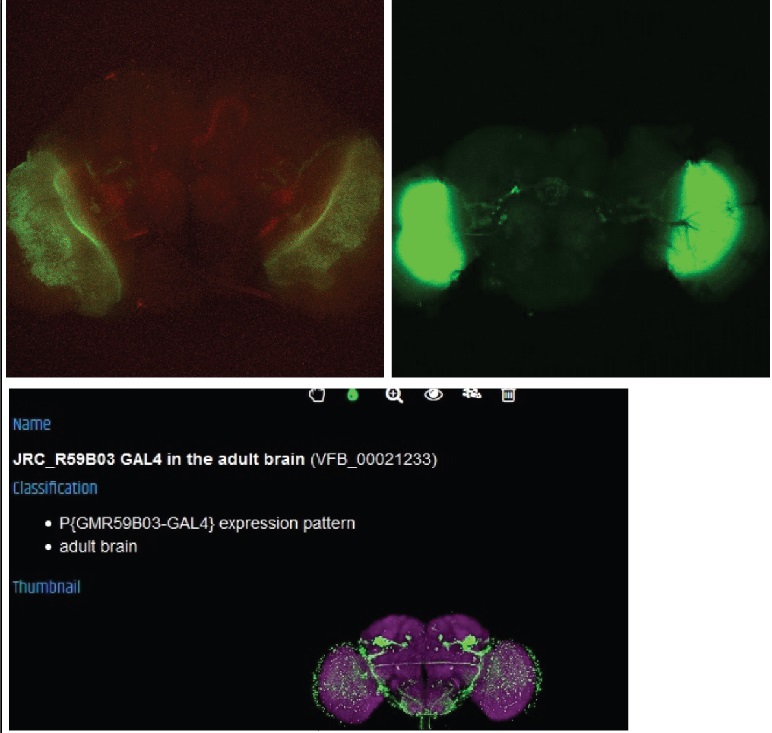
This line seems to have coexpression but the pattern also looks a bit odd, this may be due to a general weak signal and a bad dissection.
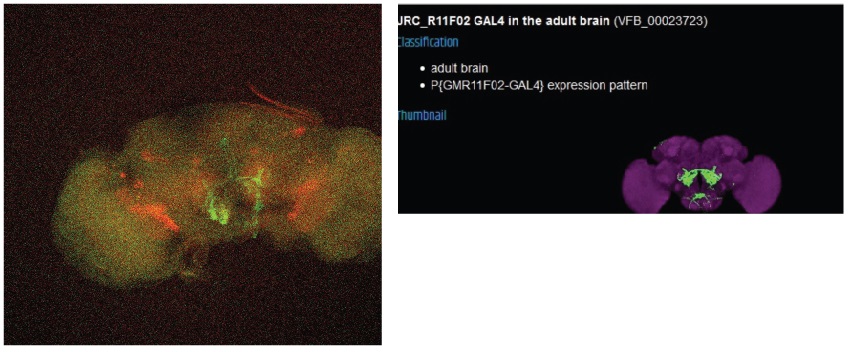
The final line seems to have no overlap with the FoxP expression.
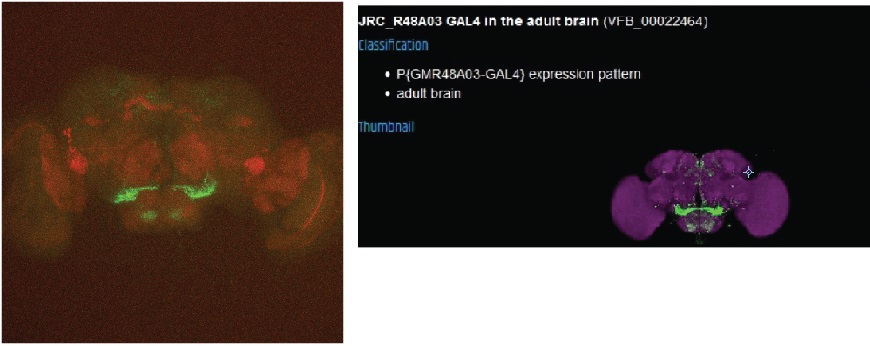
Category: Anatomy, crosses, Foxp | No Comments
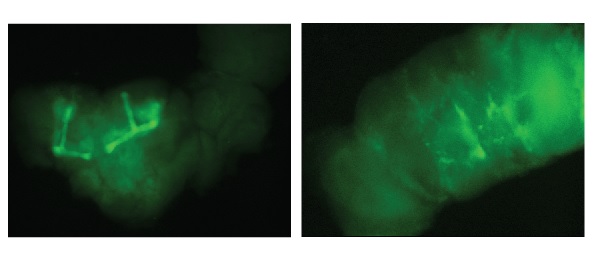
Test for hsGal4
on Monday, September 14th, 2020 1:40 | by Andreas Ehweiner
Comparison between Control (elavGal4 x CD8GFP) and hsGla4 x CD8GFP. Hs for 3h, fixation and dissection after 24h.
Category: Anatomy, crosses | No Comments
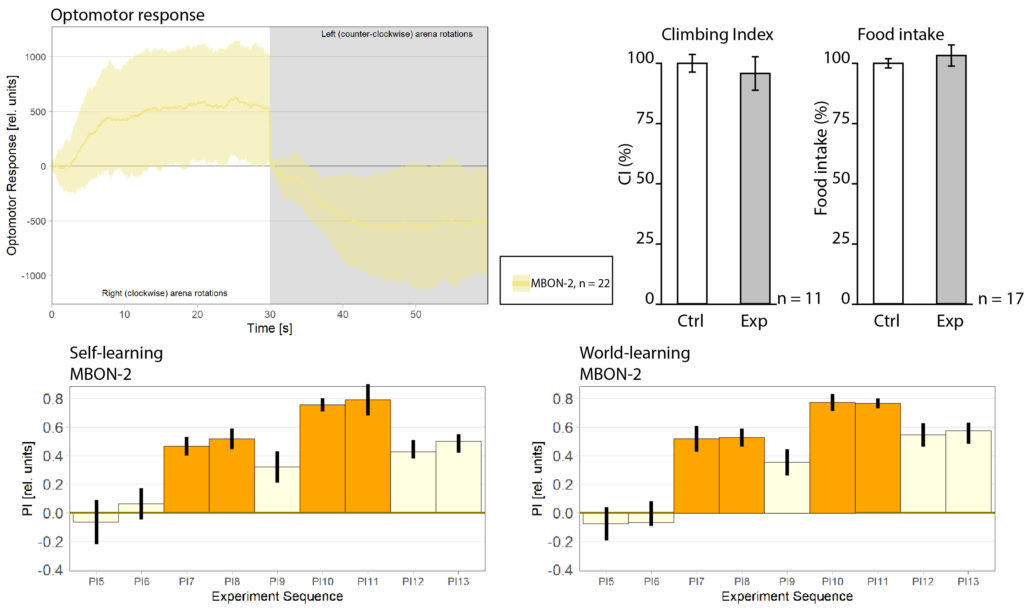
General behavior of the MBON-2
on Monday, August 31st, 2020 12:54 | by Anders Eriksson
I wanted to expand and look into some general behavior of the mbon-2 flies. Mostly as a complement to the already existing data.
Category: genetics, Lab, Memory, Operant learning, operant self-learning, Optomotor response, science | No Comments
Progress Week 29
on Monday, July 20th, 2020 1:38 | by Anders Eriksson
-Introduced Sayani to the wonderful world of Drosophila
-Been doing some DTS coding
-Preparing flies to do optomotor response for Mathias Raß

Category: flight, genetics, Lab, Memory, Optomotor response, R code | No Comments
