Bachelor Blog / #5 no learning :(
on Monday, September 11th, 2023 12:21 | by Ellie
Below you find the data from my experimental rounds A and B:
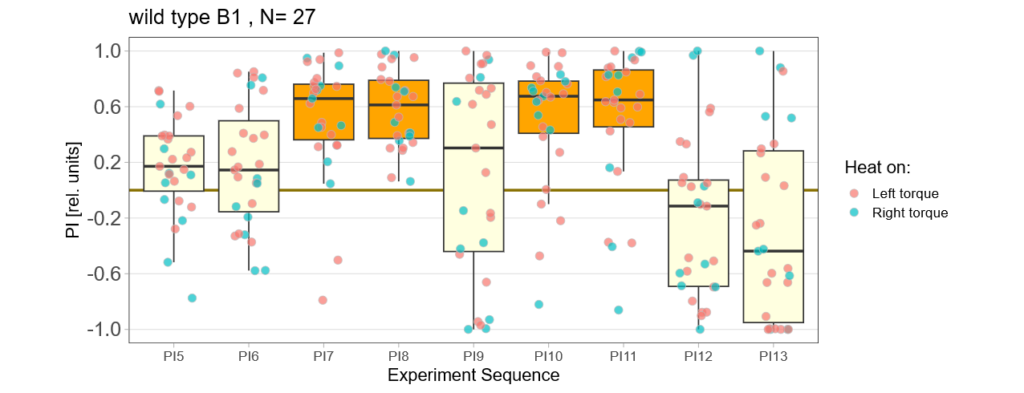
-> learning scores of the parental flies from experimental round A and B
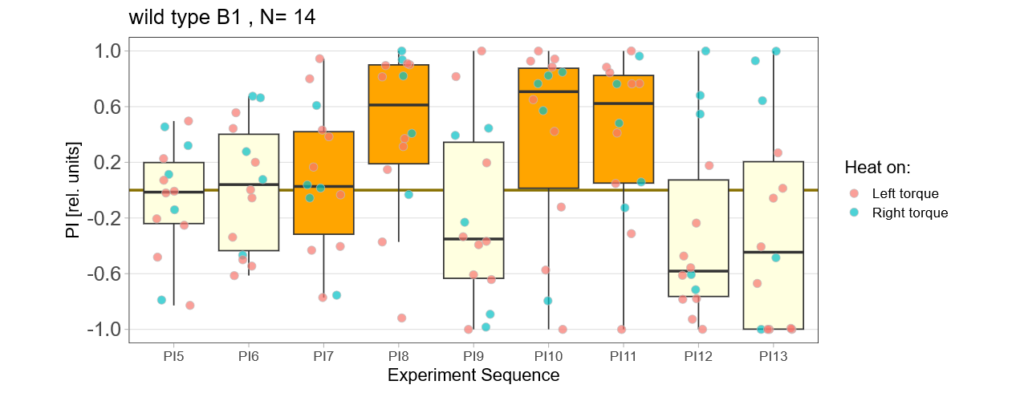
-> learning scores of the trained parent´s offspring only from experimental round A
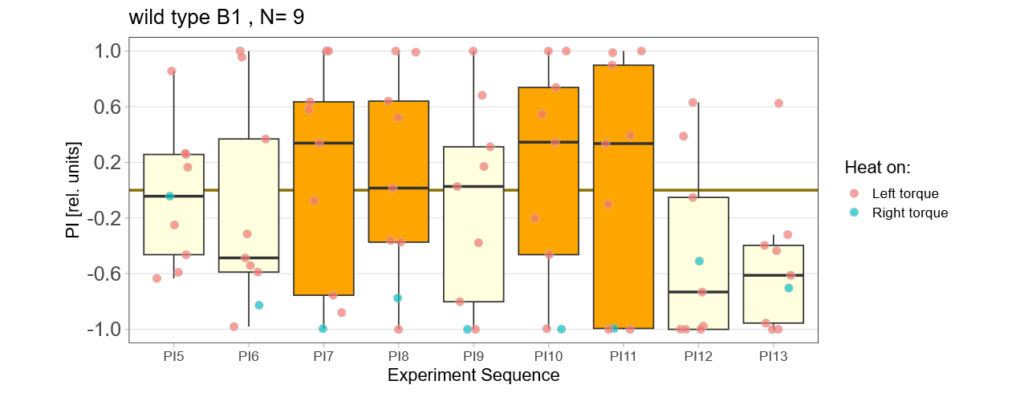
-> learning scores of the untrained parent´s offspring only from experimental round A
The results confuse me a lot and I am happy to discuss reasons :) However the offspring of the round-B will be ready for testing by the end of this week so there is still some data to collect…
Success: rsh Stock has rsh1 Mutation
on Monday, August 7th, 2023 11:11 | by Isabel Stark
Via gDNA analysis and PCR was the specific area of the rsh gene extracted and amplified where the nucleotide substitution: C to T (Folkers et al., 2006) should be for the rsh1 mutation. The amplicon was Sanger sequenced which proved the nucleotide substitution.
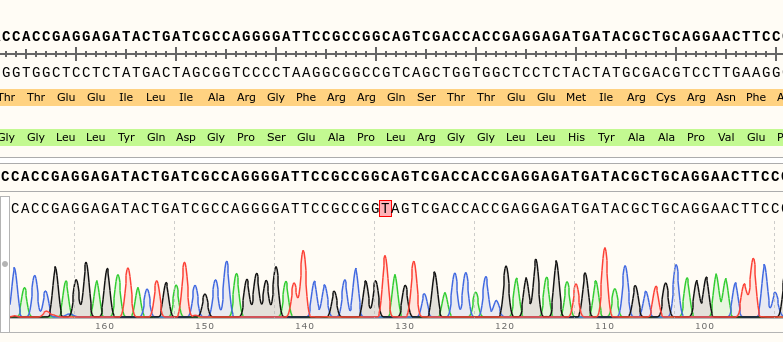
Category: genetics, Memory, Operant learning, operant self-learning, Radish, Uncategorized | No Comments
Creating gRNAs via PCR
on Friday, July 7th, 2023 6:38 | by Isabel Stark
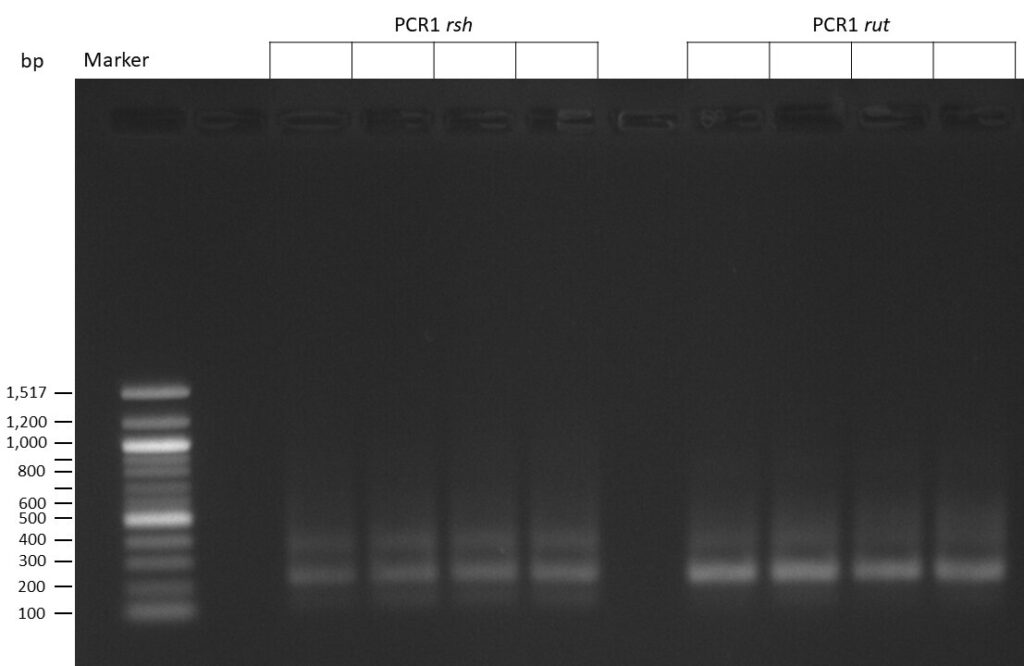
The template pCFD6 was used with a concentration of 640 pg/µl.
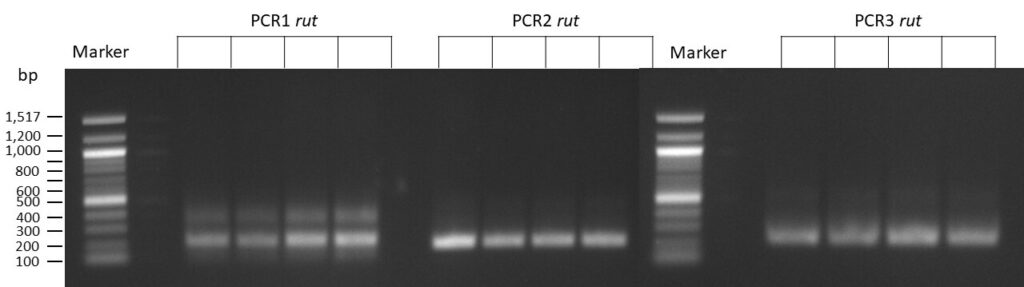
The template pCFD6 was used with a concentration of 64 pg/µl (1:10 dilution).
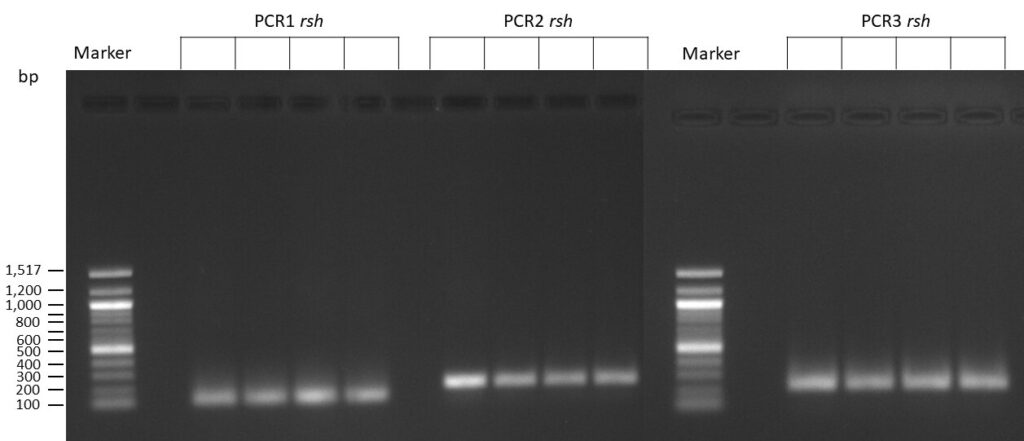
The template pCFD6 was used with a concentration of 64 pg/µl (1:10 dilution).
50µl of 5xQ5 High GC Enhancer was added to the PCR mix.
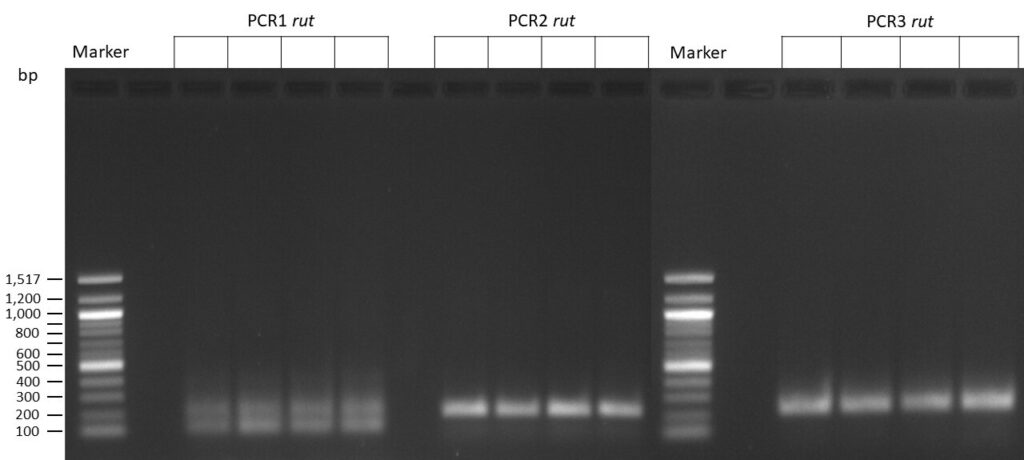
The template pCFD6 was used with a concentration of 128 pg/µl (1:5 dilution).
50µl of 5xQ5 High GC Enhancer was added to the PCR mix.
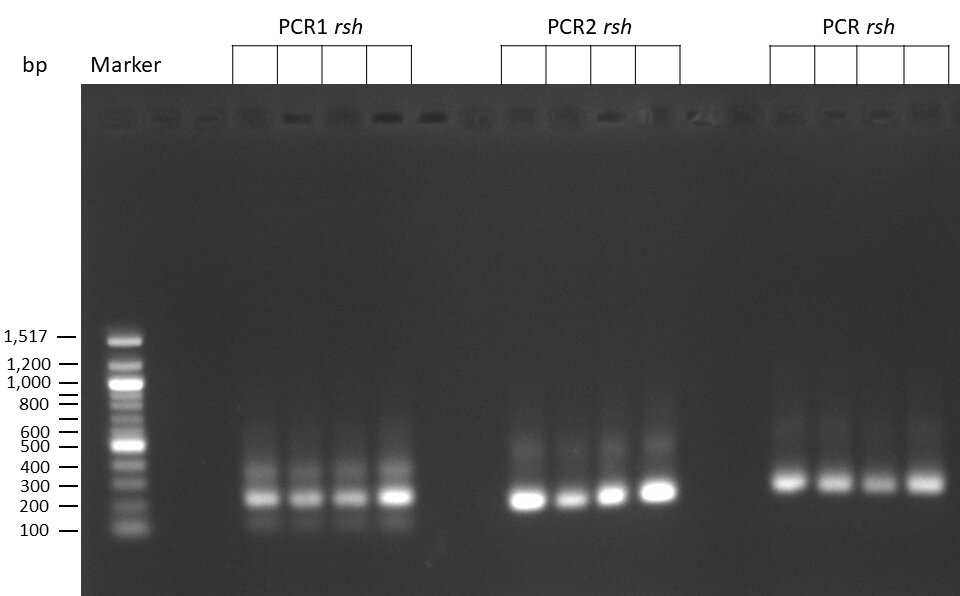
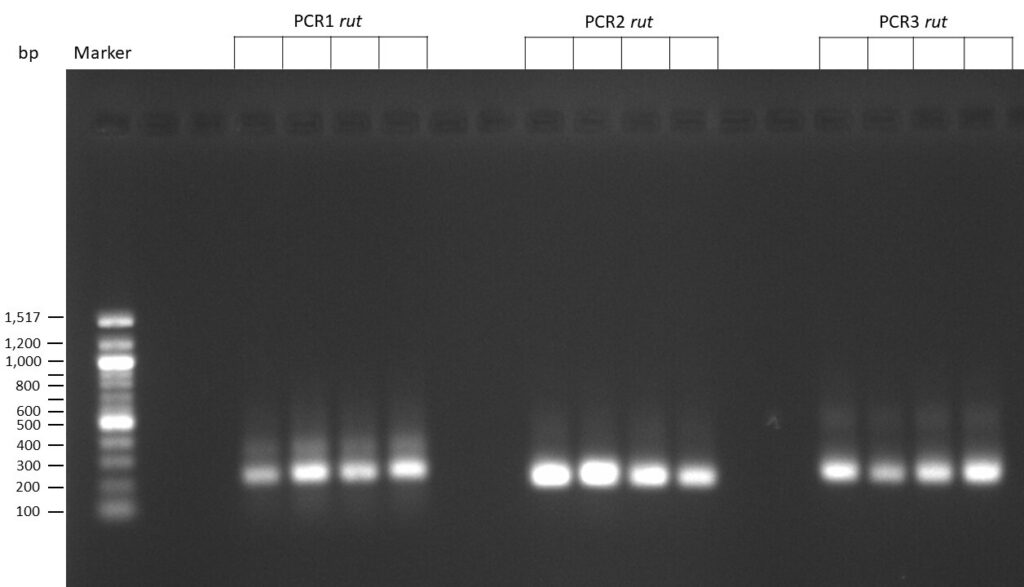
The template pCFD6 was used with a concentration of 640 pg/µl.
The Phusion DNA Polymerase was used instead of the Q5 High-Fidelity DNA Polymerase.
Category: genetics, Memory, Operant learning, operant self-learning, Radish, Uncategorized | No Comments
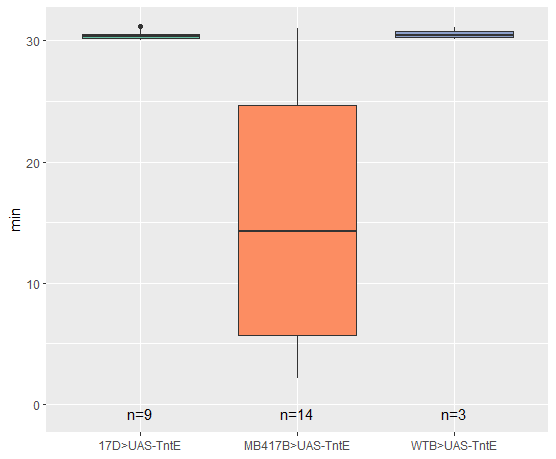
Flight Duration
on Monday, January 30th, 2023 12:40 | by Silvia Marcato
Category: Uncategorized | No Comments
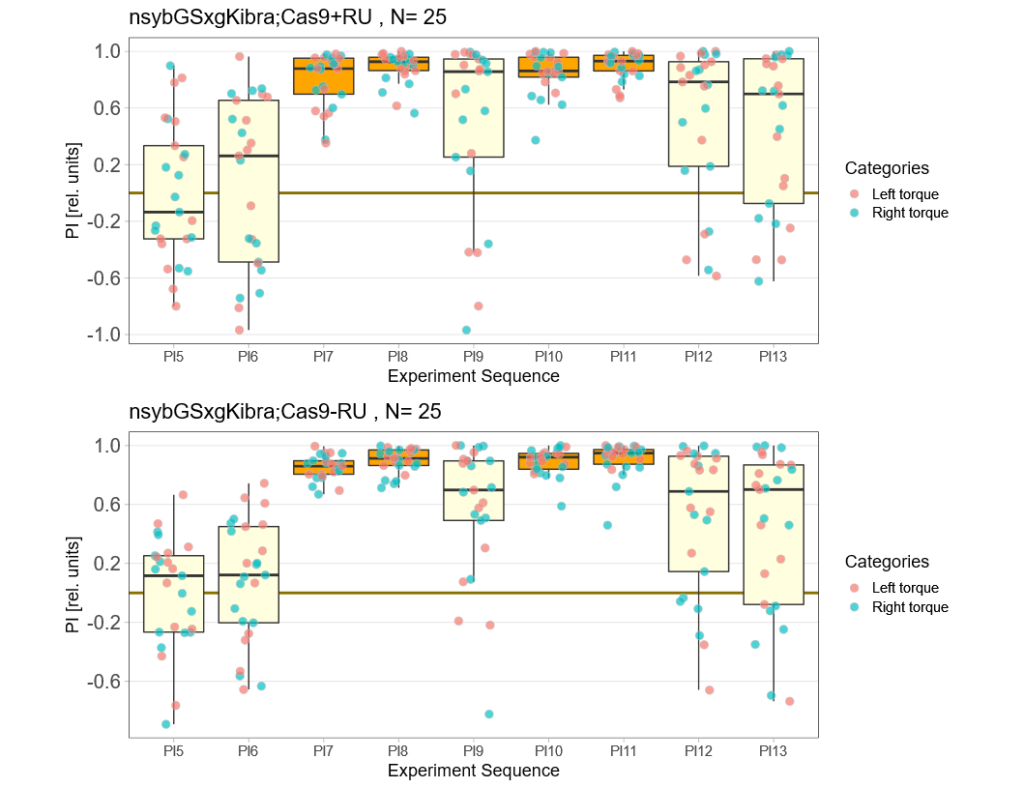
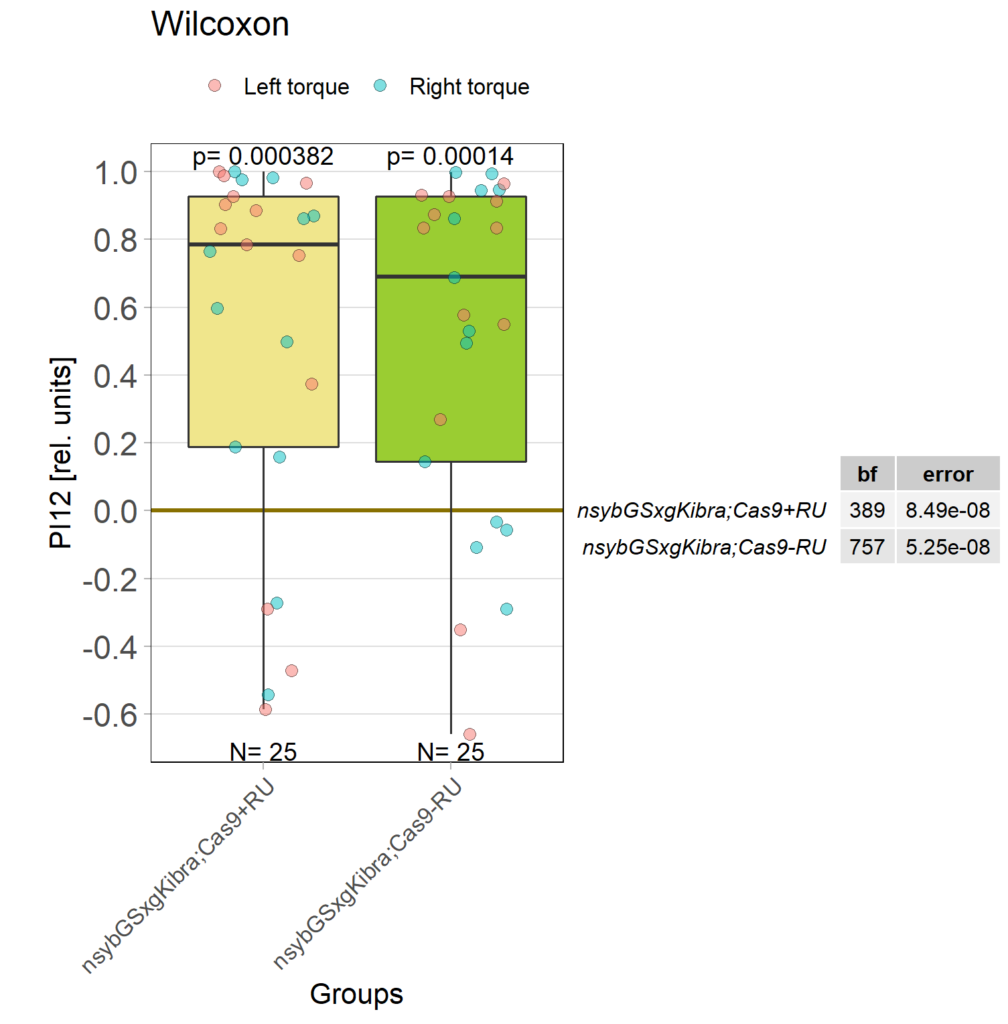
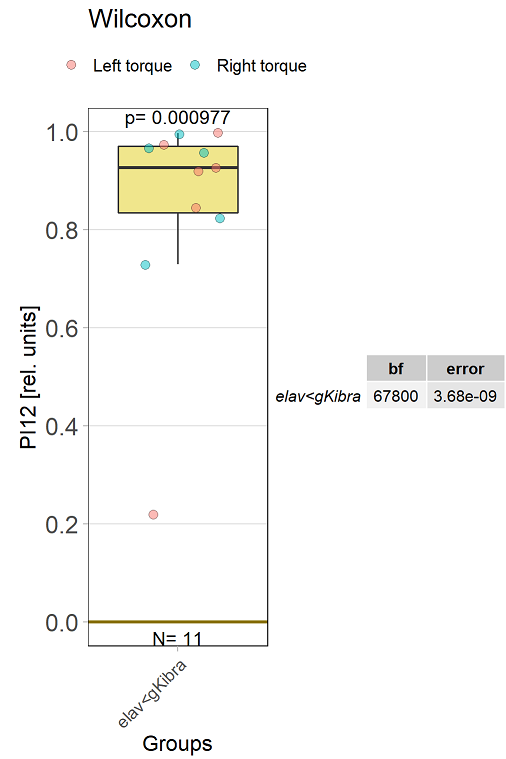
Kibra knockout, adult
on Monday, December 19th, 2022 11:15 | by Andreas Ehweiner
Category: flight, Memory, Operant learning, operant self-learning, Uncategorized | No Comments
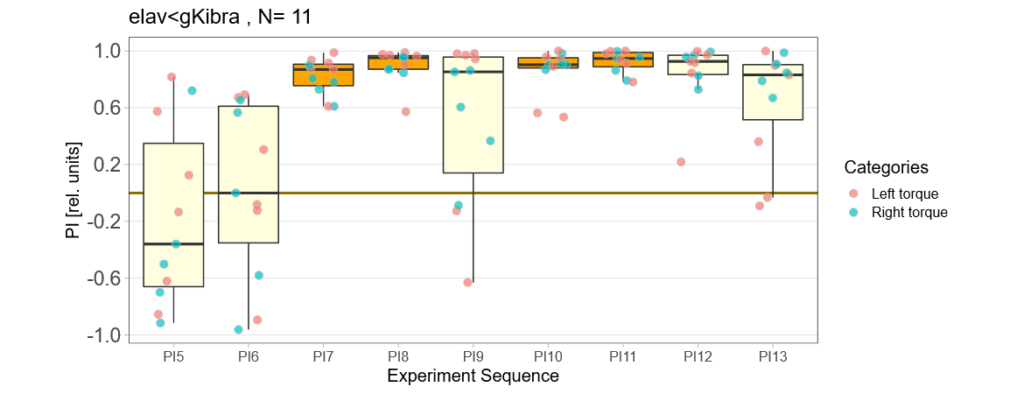
Kibra developmental knockout
on Monday, September 5th, 2022 9:39 | by Andreas Ehweiner
Category: flight, Memory, Operant learning, operant self-learning, Uncategorized | No Comments
Enes and Aslıhan joystick results of ATR and NOATR Control, ATR SifaG4 and ATR 13.0273 for yellow light.
on Wednesday, August 24th, 2022 10:32 | by Enes Seker
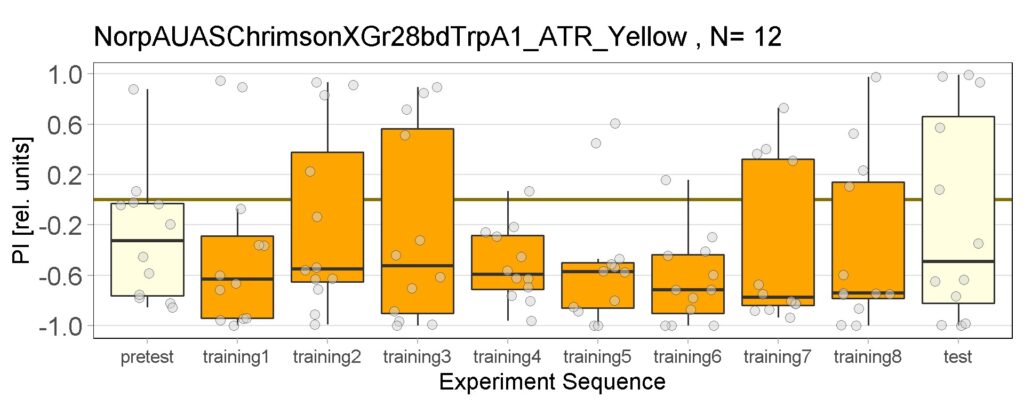
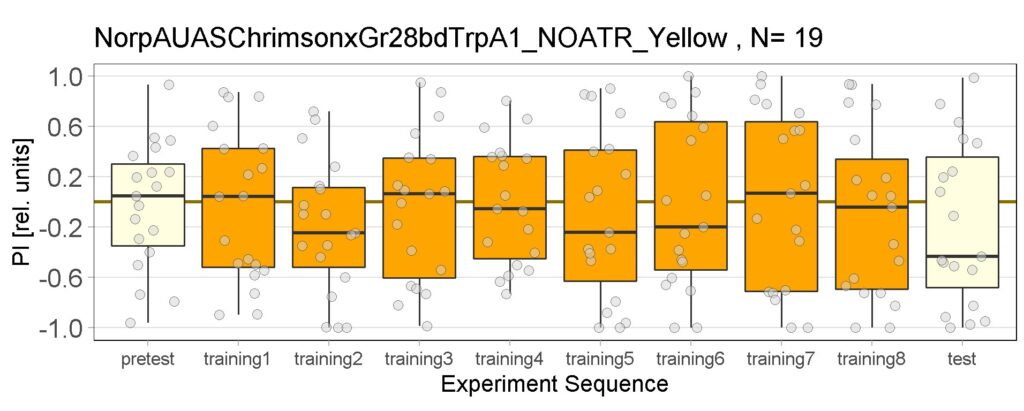
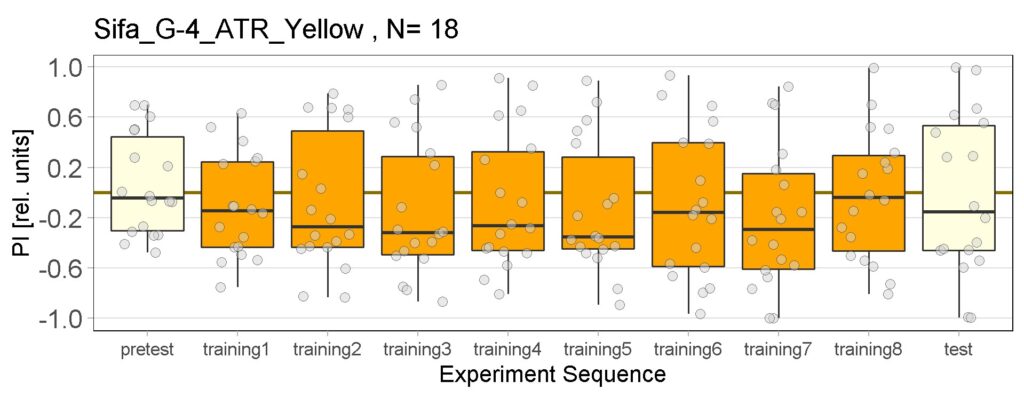
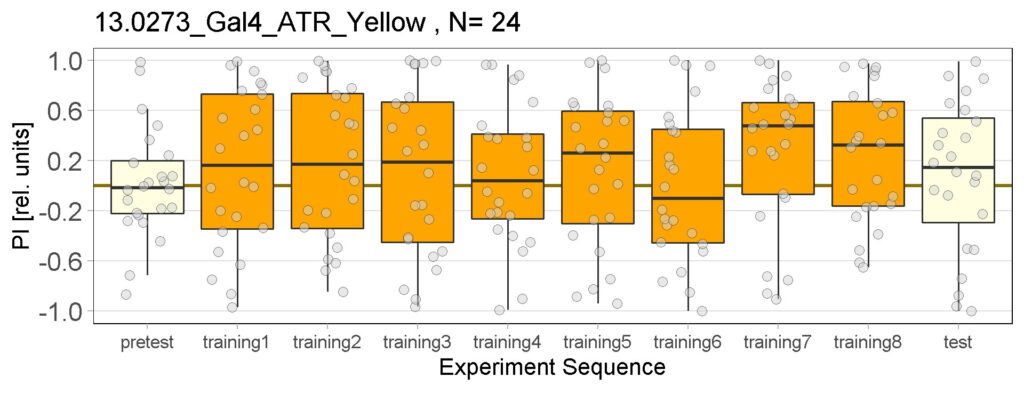
New experiments added for yellow light, ATR and NOATR Control group, ATR Sifa-G4 and ATR 13.0273-Gal4.
Category: Optogenetics, Uncategorized | No Comments
Enes and Aslıhan Joystick Experiment of Control groups , Sifa-G4, 13.0273-Gal4
on Monday, August 22nd, 2022 11:48 | by Enes Seker
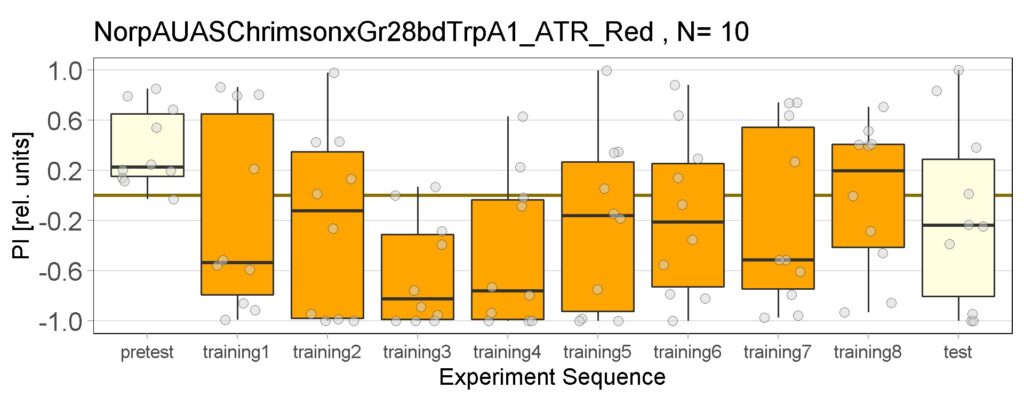
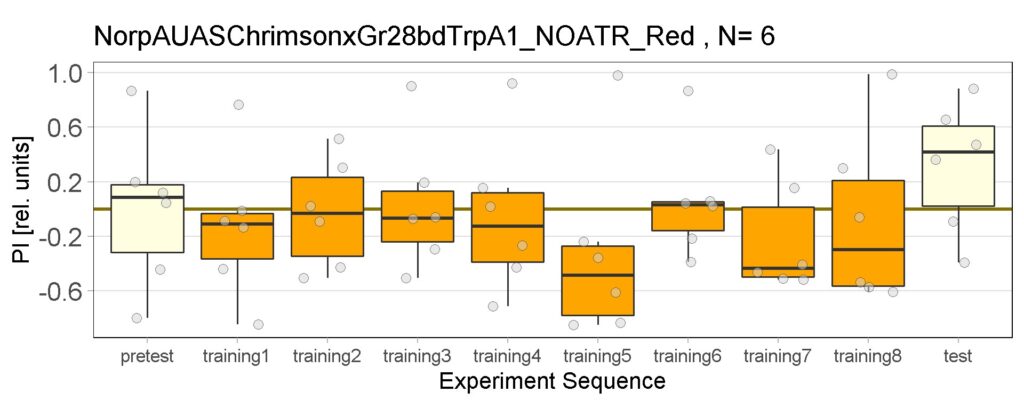
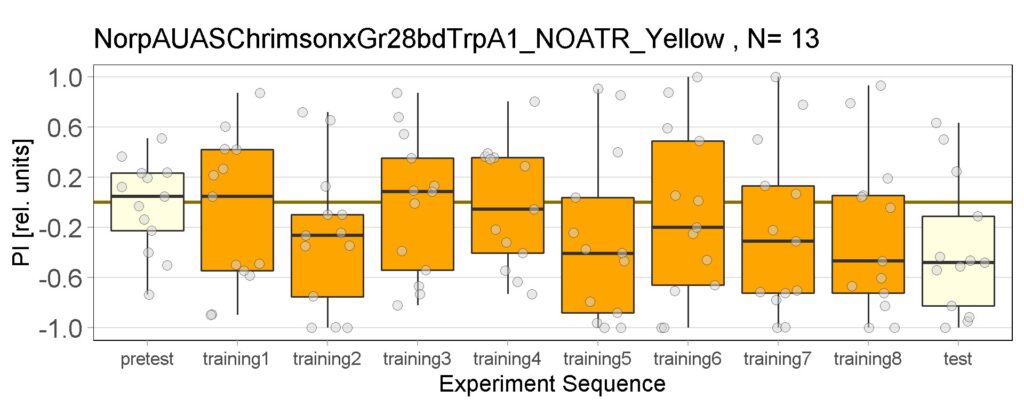
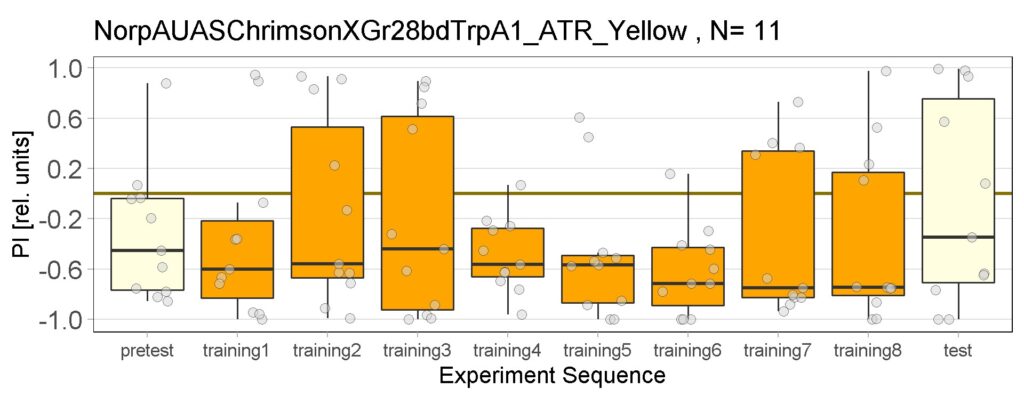
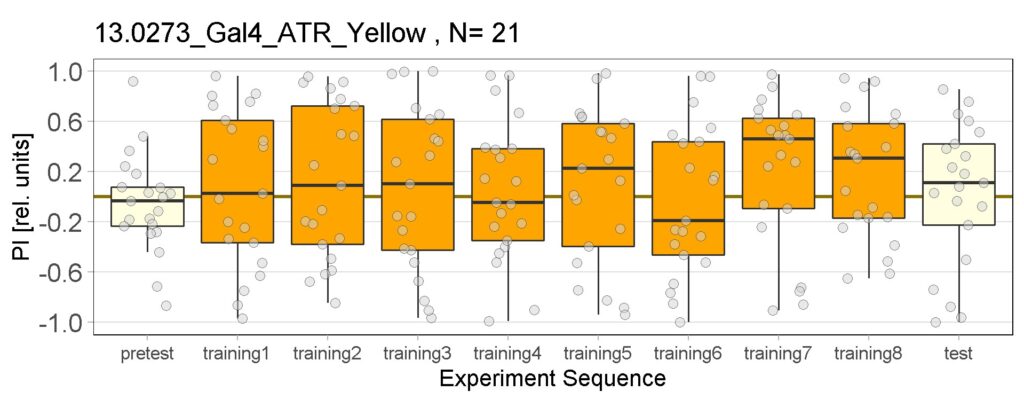
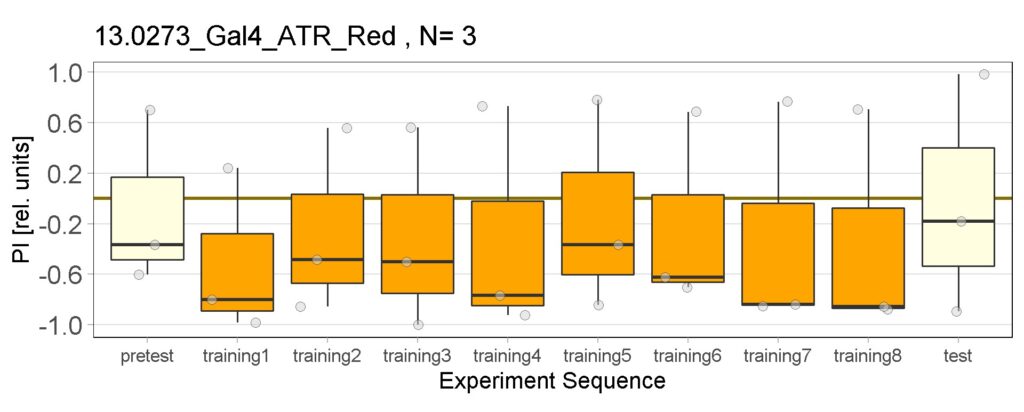
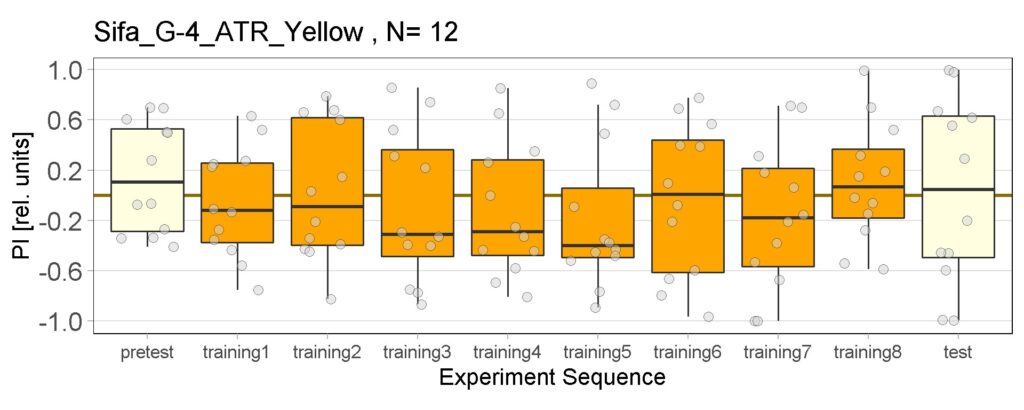
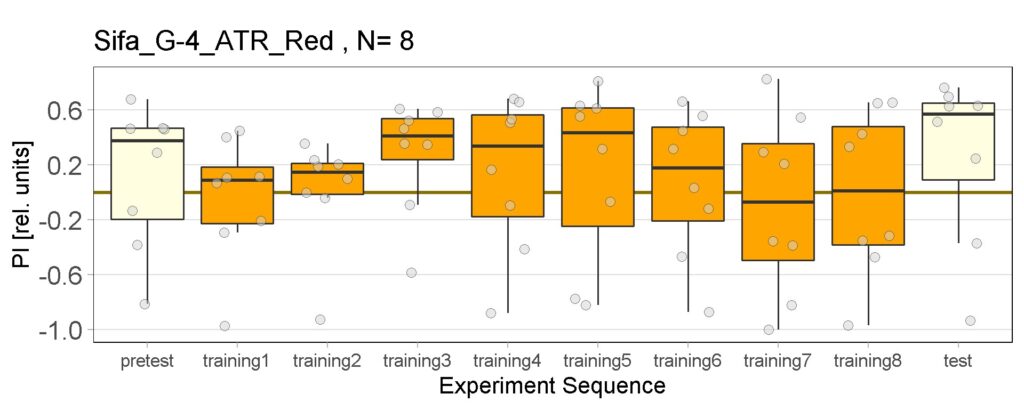
These are the result of Joystick experiements of Control group with ATR and without ATR yellow and red light, SifaG4 group with ATR, yellow and red light, 13.0273Gal4 group with ATR, yellow and red light.
Category: Uncategorized | No Comments
Enes & Aslı & Melis Control Groups
on Thursday, August 18th, 2022 10:38 | by Melis Öztibet
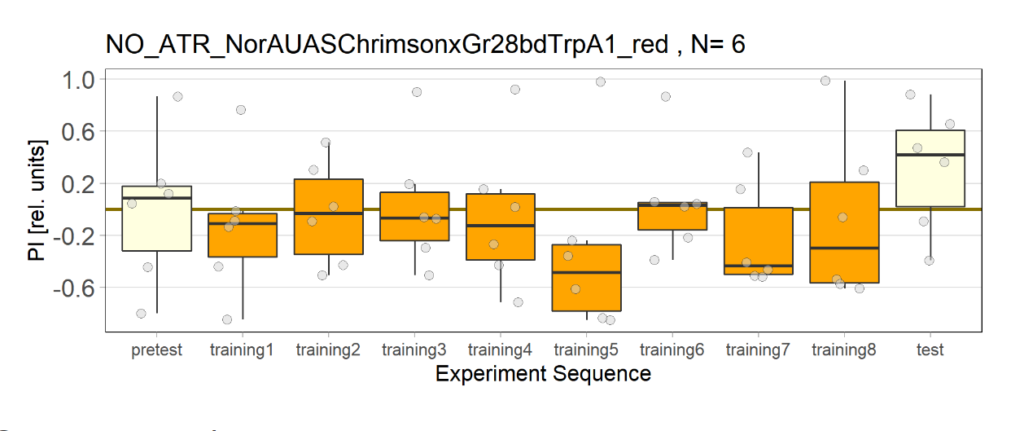

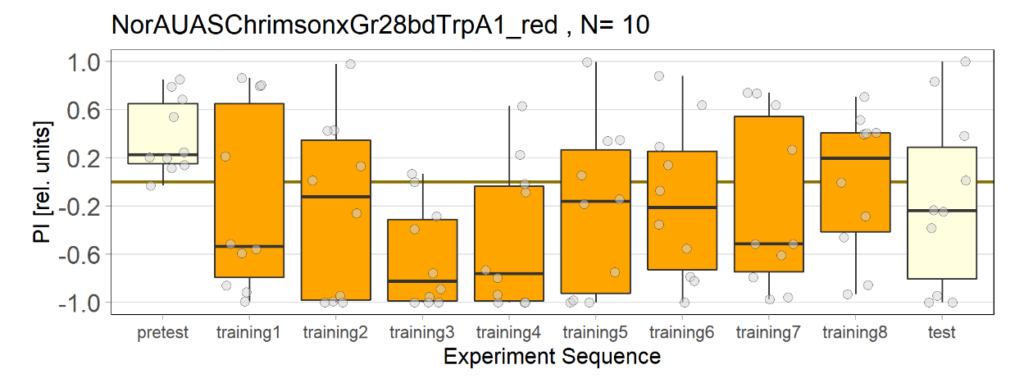
Category: Uncategorized | No Comments
NorAUASChrimsonxGr28bdTrpA1_Red
on Monday, July 18th, 2022 9:15 | by Melis Öztibet
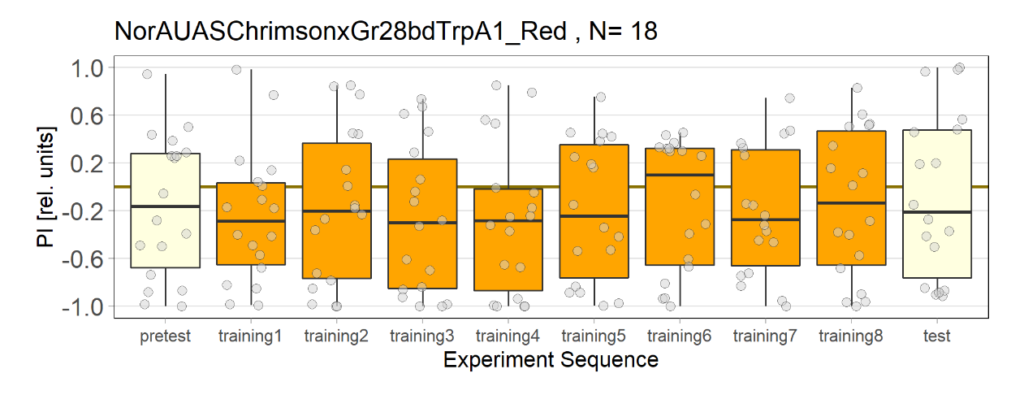
Category: Uncategorized | No Comments