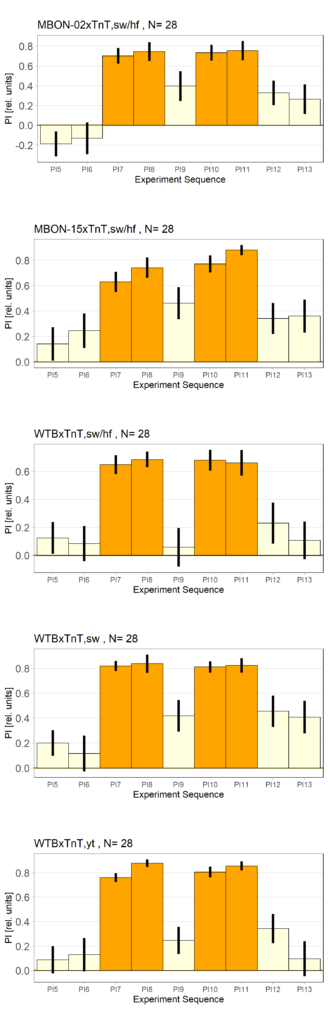
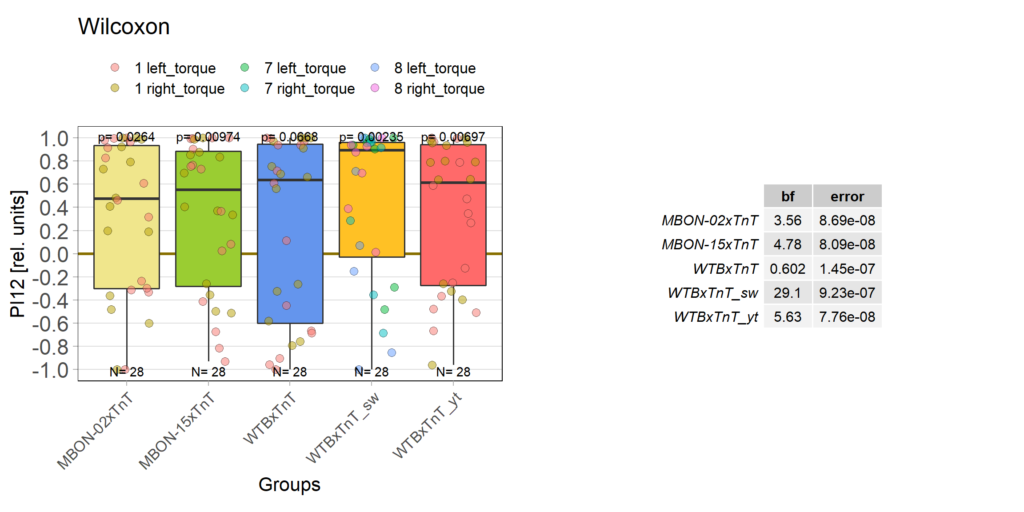
More data from the basement
on Monday, July 8th, 2024 8:15 | by Ellie
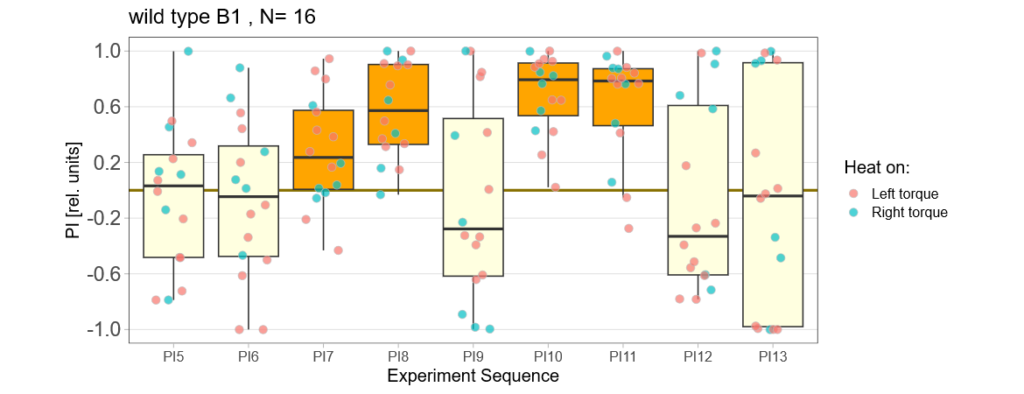
With the modified laser settings I was able to test 3 more wtb. Here are the results:

–> yaw torque
Testing the basement flight simulator
on Wednesday, June 19th, 2024 3:46 | by Ellie
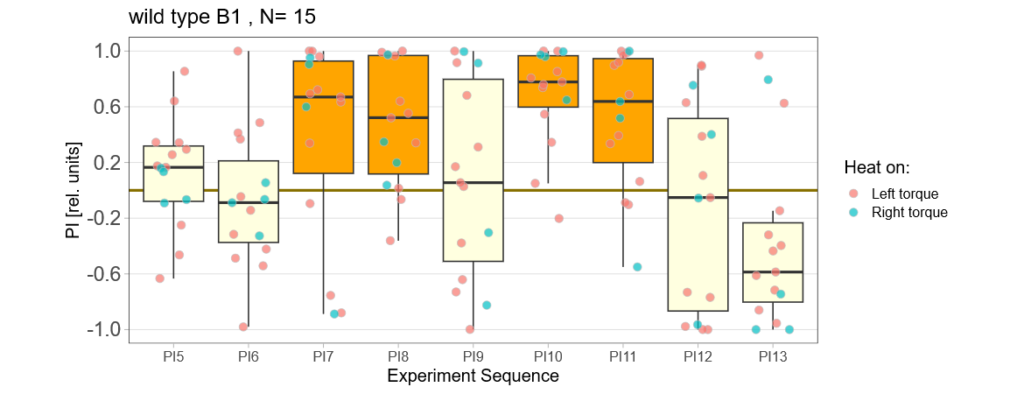
Before collecting the actual data I make sure that the flight simulator in the basement does its thing ;) Here are the test runs with wtb flies for different protocols:

—>> yaw torque

–>> switch mode

–>> habit formation
Final ‘Joystick’ test results
on Monday, January 29th, 2024 11:30 | by Devashree Joglekar
Over the course, control and test lines were experimented with under ‘Joystick’. The test lines include 13:0273-Gal 4 line, SS56699 line, and TH-C-AD; TH-D-DBD line. The tests were done under red and yellow light conditions.
Red light conditions:
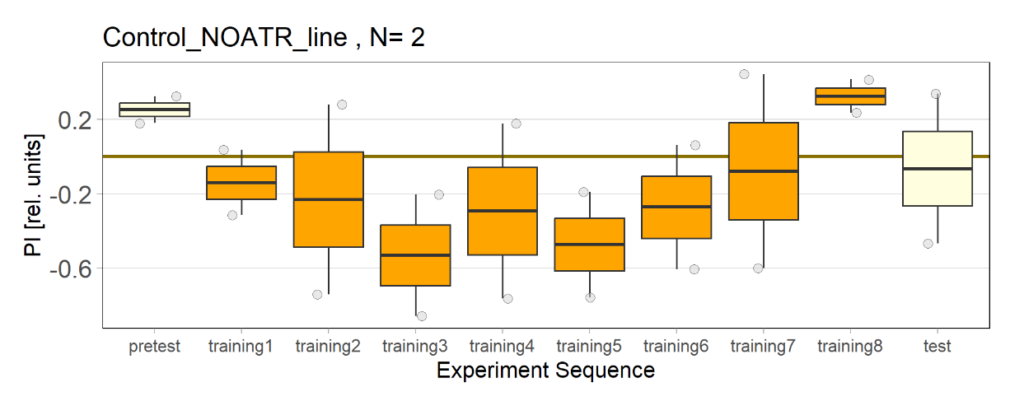
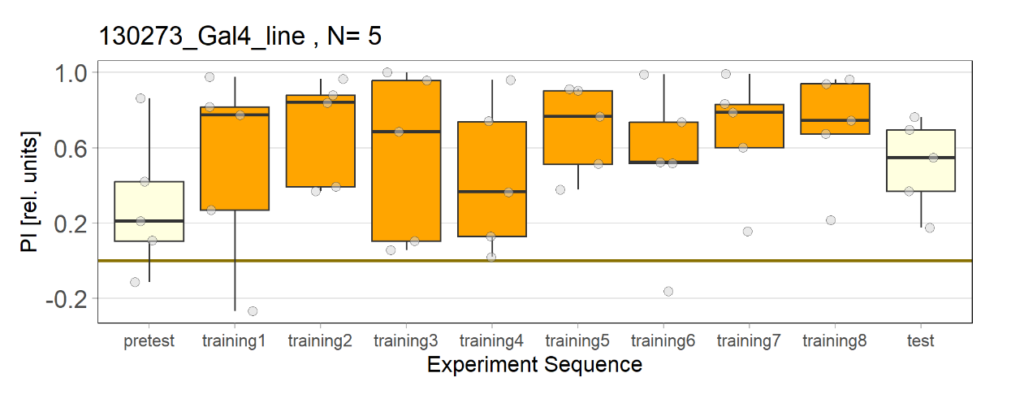
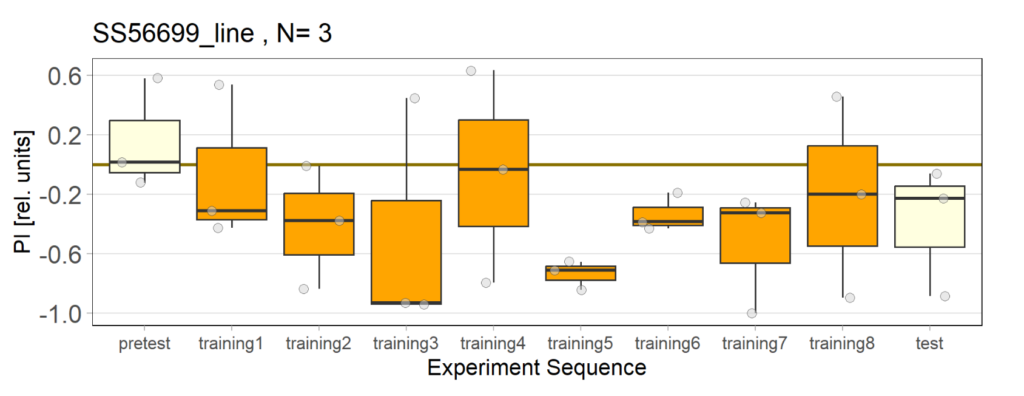
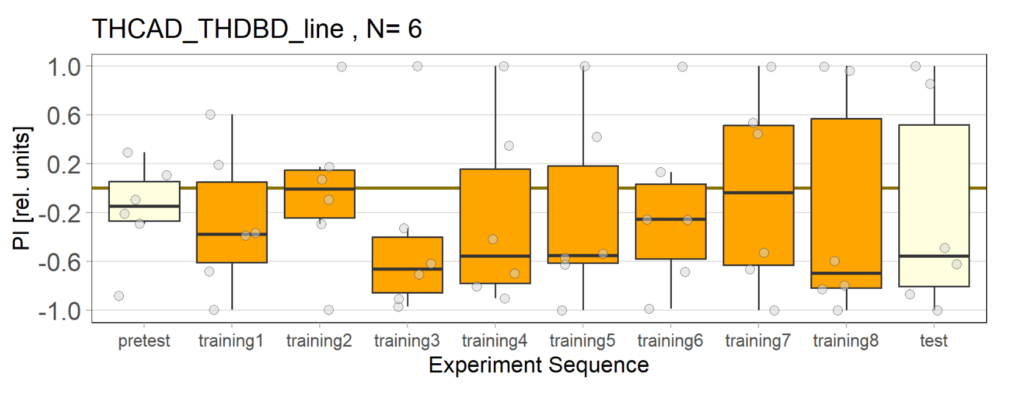
Yellow light conditions:
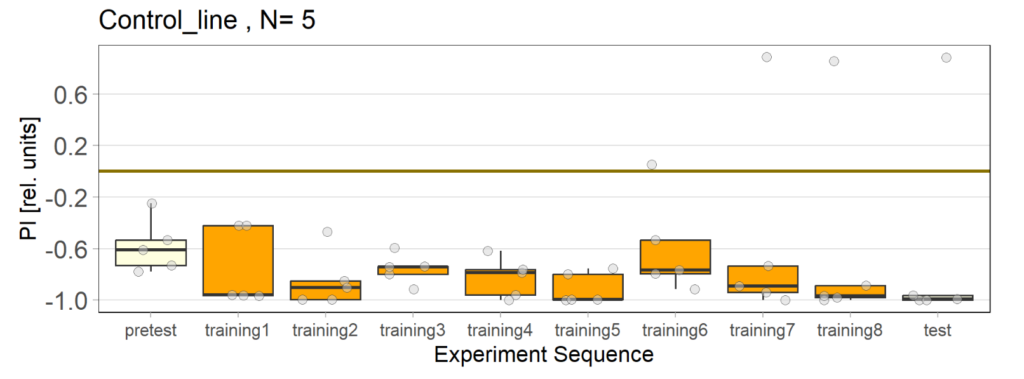
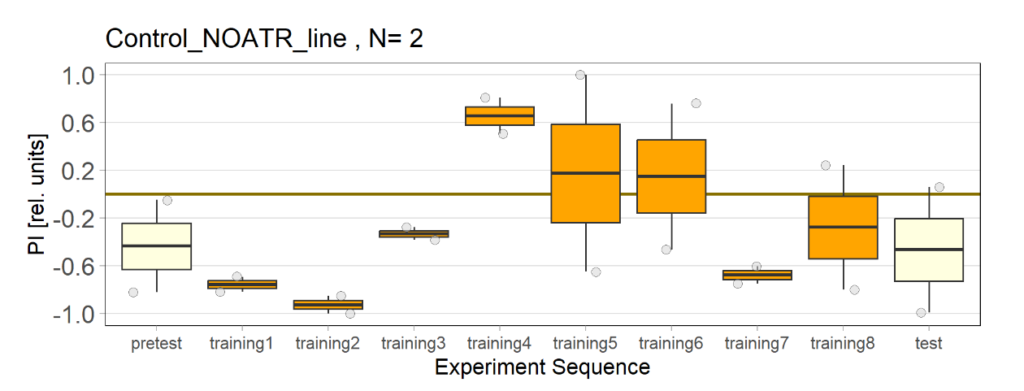
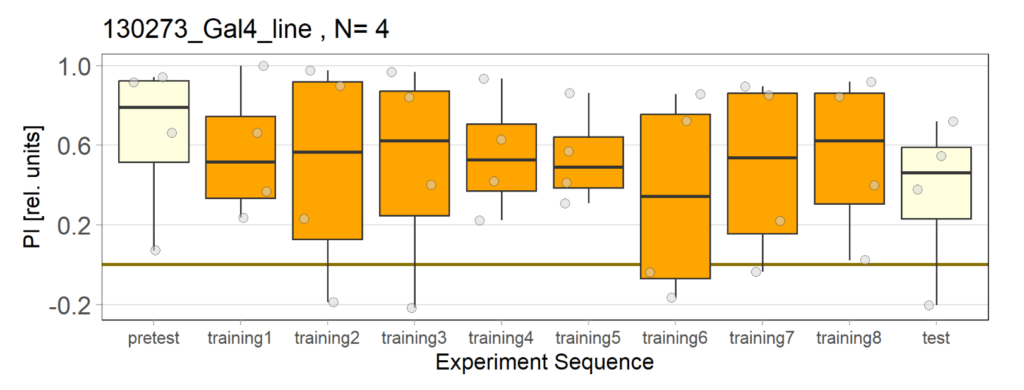
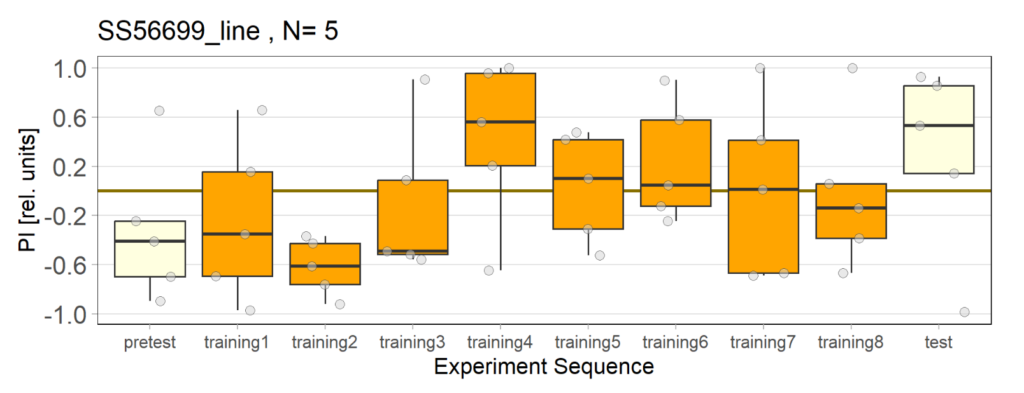
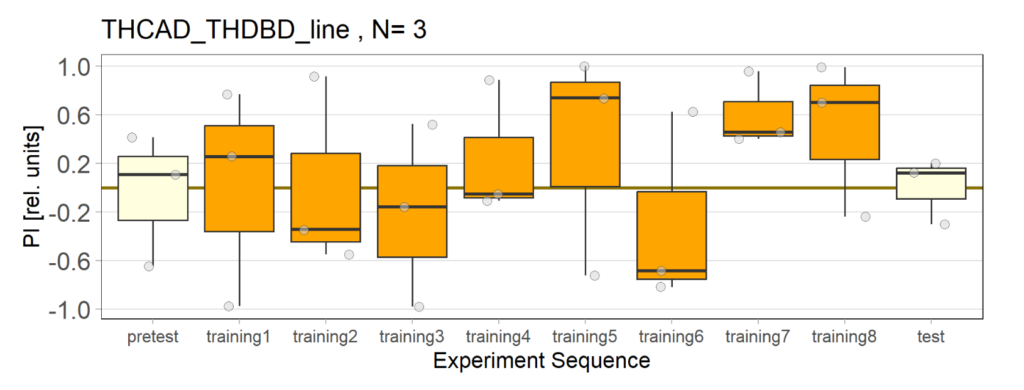
Category: Operant reinforcment, Optogenetics | No Comments
Bachelor Blog / #7 offspring
on Monday, October 2nd, 2023 10:51 | by Ellie
Below you can find the data I collected from the offspring flies:
-> offspring from trained parents
-> offspring from untrained parents
(I left out data from flies that showed negative preference during two training periods in a row)
Bachelor Blog / #6 playing around
on Monday, September 18th, 2023 12:51 | by Ellie
Since the results I got after training the parental flies looked a bit odd on first sight I decided to take a closer view…
First I excluded some weird animals that either showed a larger preference for one side than avoidance or showed no avoidance two training periods in a row:
Next I compared the behavior of flies that showed avoidance but no learning with the behavior of the remaining flies:
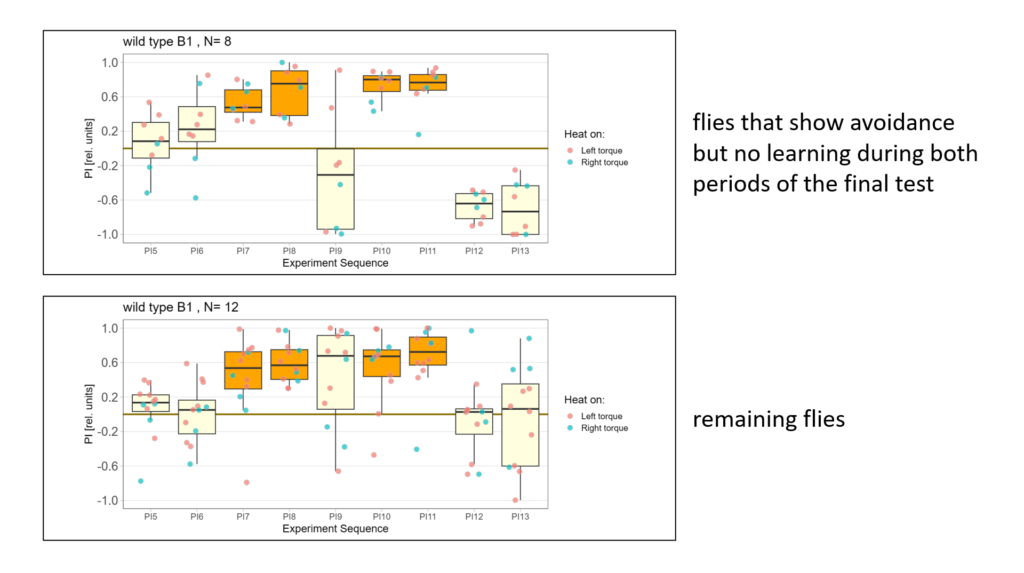
-> Avoidance is almost the same but note the first test period!
Lastly I split the data according to male and female flies. Here is what I got:
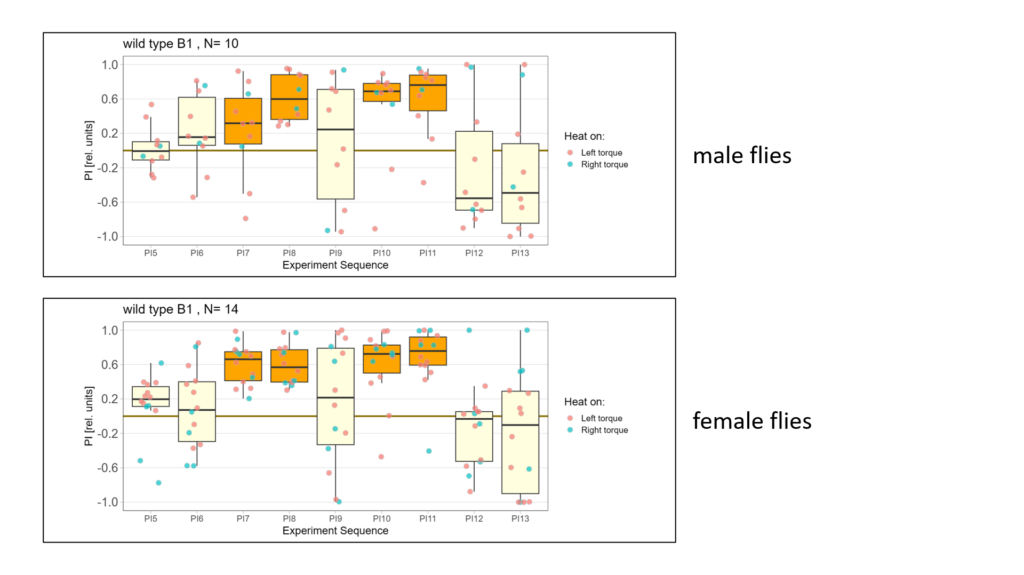
-> Looks a bit like there is negative learning in the male flies however I don´t have enough data to be sure…
As an overview here are all the flies (except for the excluded ones) together again:
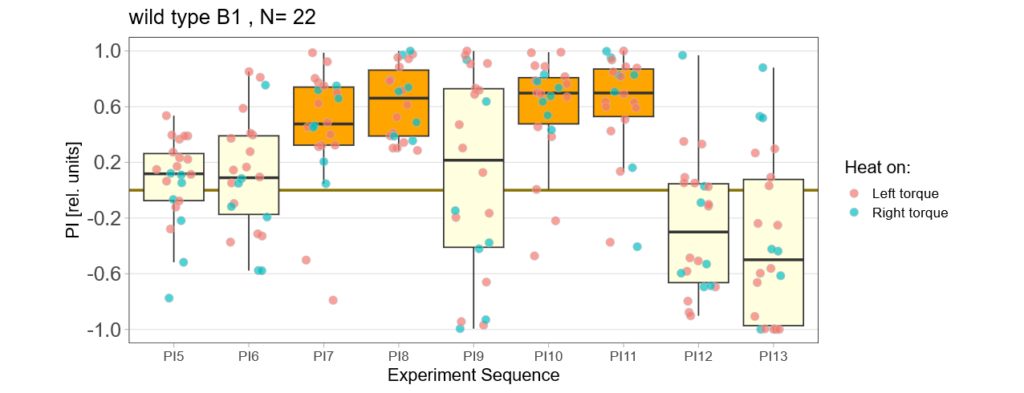
Bachelor Blog / #5 no learning :(
on Monday, September 11th, 2023 12:21 | by Ellie
Below you find the data from my experimental rounds A and B:
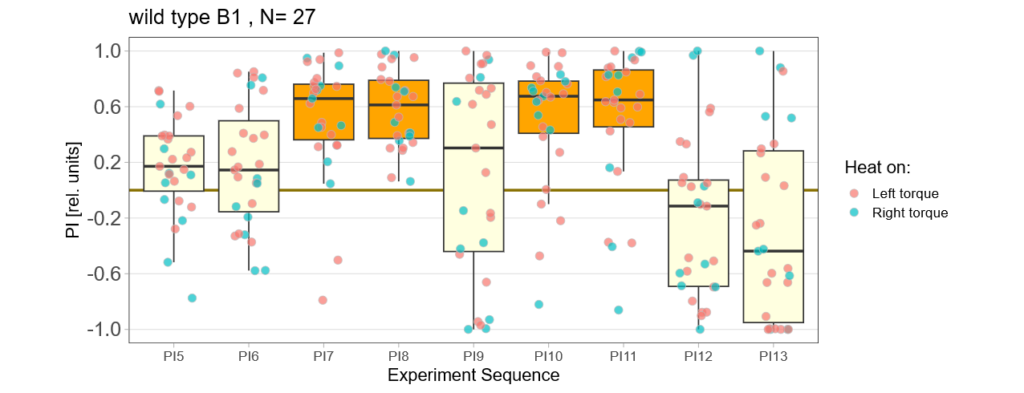
-> learning scores of the parental flies from experimental round A and B
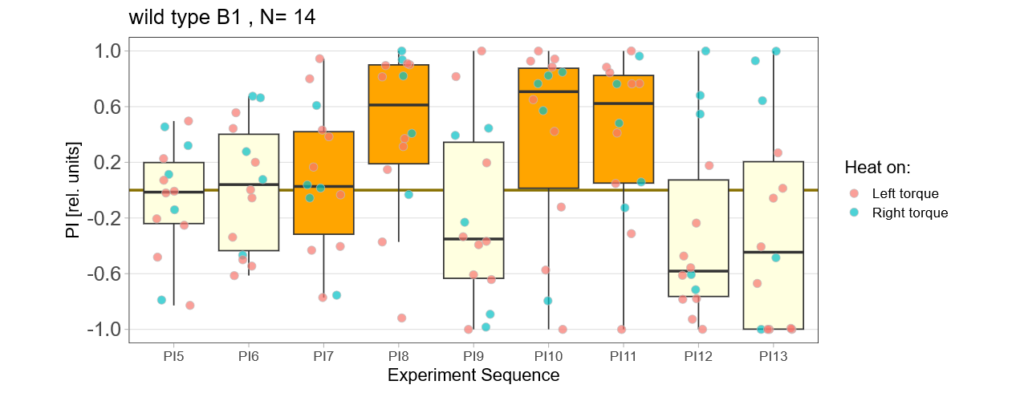
-> learning scores of the trained parent´s offspring only from experimental round A
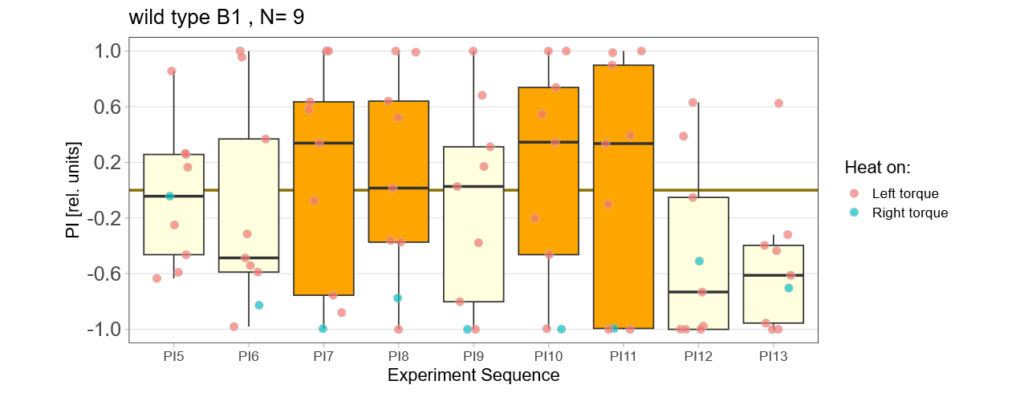
-> learning scores of the untrained parent´s offspring only from experimental round A
The results confuse me a lot and I am happy to discuss reasons :) However the offspring of the round-B will be ready for testing by the end of this week so there is still some data to collect…
MBON screen verification
on Tuesday, December 21st, 2021 9:04 | by Radostina Lyutova
Category: MBON, Memory, Operant learning, Operant reinforcment, operant self-learning, World learning | No Comments
World- vs. self-learning, MBON screen verification, Laser 3.06V
on Monday, November 29th, 2021 1:14 | by Radostina Lyutova
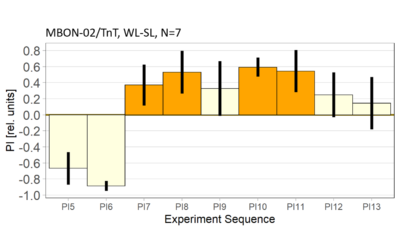
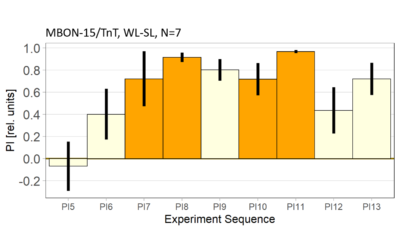
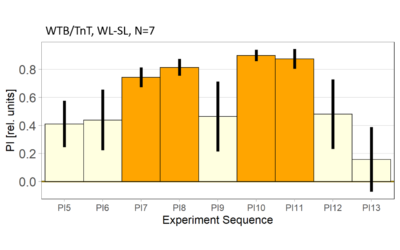
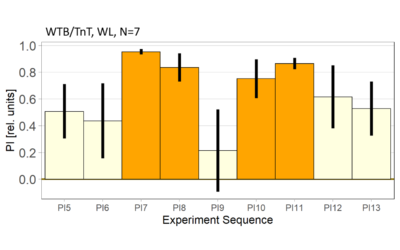
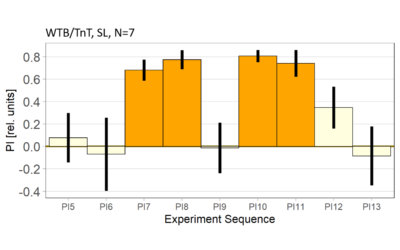
Category: flight, MBON, Memory, Operant learning, Operant reinforcment, operant self-learning, World learning | No Comments
New batch of ATR wrecks screen results?
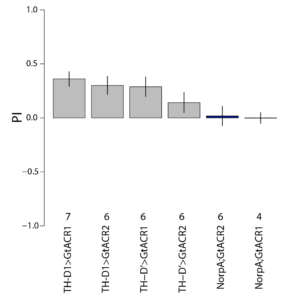
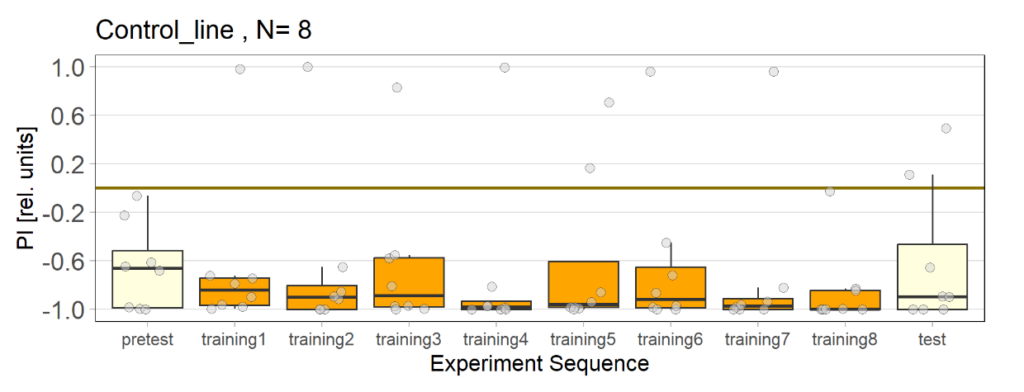
on Monday, May 4th, 2020 1:16 | by Björn Brembs
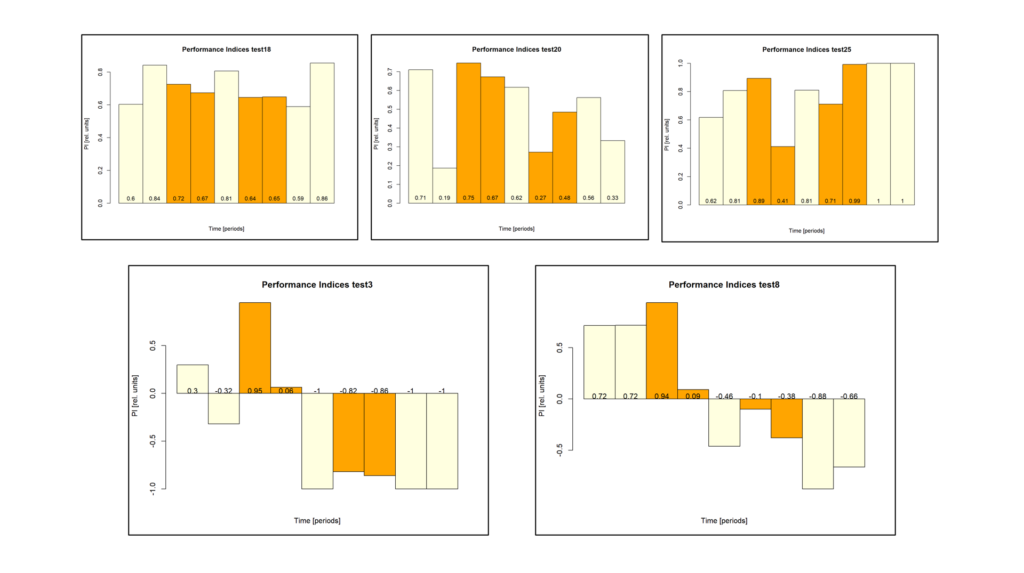
At the beginning of last week, the flies in the optogenetics rescreen seemed to behave very different compared to all the weeks before: groups that kept the lights on now switched it off and vice versa. There was also some of that in the control flies, but to a lesser extent. This is what the last training PIs looked like for the seven groups before last week:
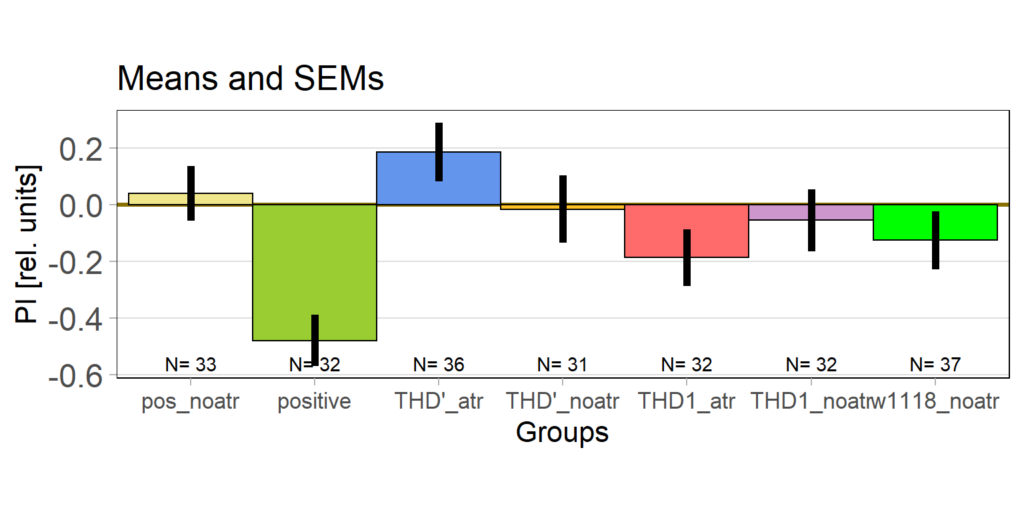
The data from last week then looked like this:
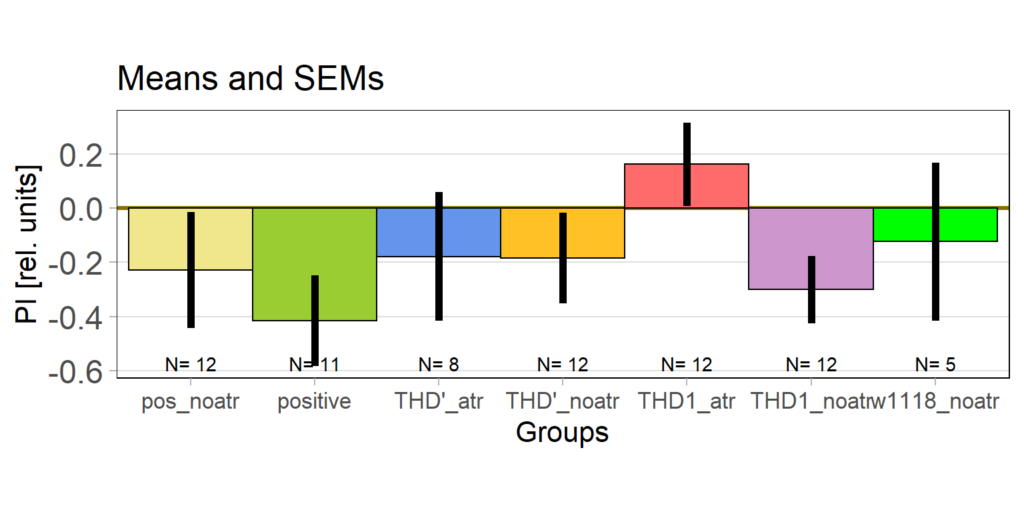
Especially the two groups that did receive ATR reversed their previous screen results. Potentially, reduced concentration of ATR may have reduced the effect of light which may have led to the reversal of the effects.
Category: neuronal activation, Operant reinforcment, Optogenetics | No Comments
Inhibiting the interesting lines
on Monday, October 8th, 2018 2:44 | by Christian Rohrsen
Category: neuronal activation, Operant reinforcment, Optogenetics | No Comments