Small but important differences
on Monday, July 22nd, 2024 8:52 | by Björn Brembs
Slowly the data are filling up and we start to see some differences emerge between the controls and the aPKC knock-outs:

We still need to get to about N=40, so there is still some way to go.
Category: operant self-learning, PKC | No Comments
Quality control reduced number of animals
on Monday, July 15th, 2024 8:34 | by Björn Brembs
Going over the optomotor responses with a fine comb revealed a bunch of flies where the algorithm wasn’t able to provide a proper fit for the OMR asymptote. Therefore, I will need more time to finish the data set. Here the current torque-learning PIs:

Clearly, the genetic controls learn while the flies with knocked-out aPKC in FoxP neurons fail to show a significant learning score. However, the OMR asymmetry effect in the genetic controls appears weaker than the one we discovered in WTB flies, as can be seen in the OMR traces after the self-learning:

Then again, at the .05 level, the asymmetry index is significant. Not the alpha level we commonly use, but also a lower N than we strive for (above is before training, below is after):

The transgenic experimental flies, in contrast, don’t seem to show much of an effect at all:


Category: Foxp, operant self-learning, PKC | No Comments
Almost there
on Monday, July 8th, 2024 8:33 | by Björn Brembs
Not many fliers left now. Will start evaluating optomotor asymmetry now.

Category: operant self-learning, Optomotor response, PKC | No Comments
Yaw torque avoidance reference
on Monday, June 24th, 2024 10:01 | by Björn Brembs
Category: Foxp, Operant learning, operant self-learning, PKC | No Comments
Testing the basement flight simulator
on Wednesday, June 19th, 2024 3:46 | by Ellie
Before collecting the actual data I make sure that the flight simulator in the basement does its thing ;) Here are the test runs with wtb flies for different protocols:

—>> yaw torque

–>> switch mode

–>> habit formation
Passing the halfway mark
on Monday, June 17th, 2024 8:25 | by Björn Brembs
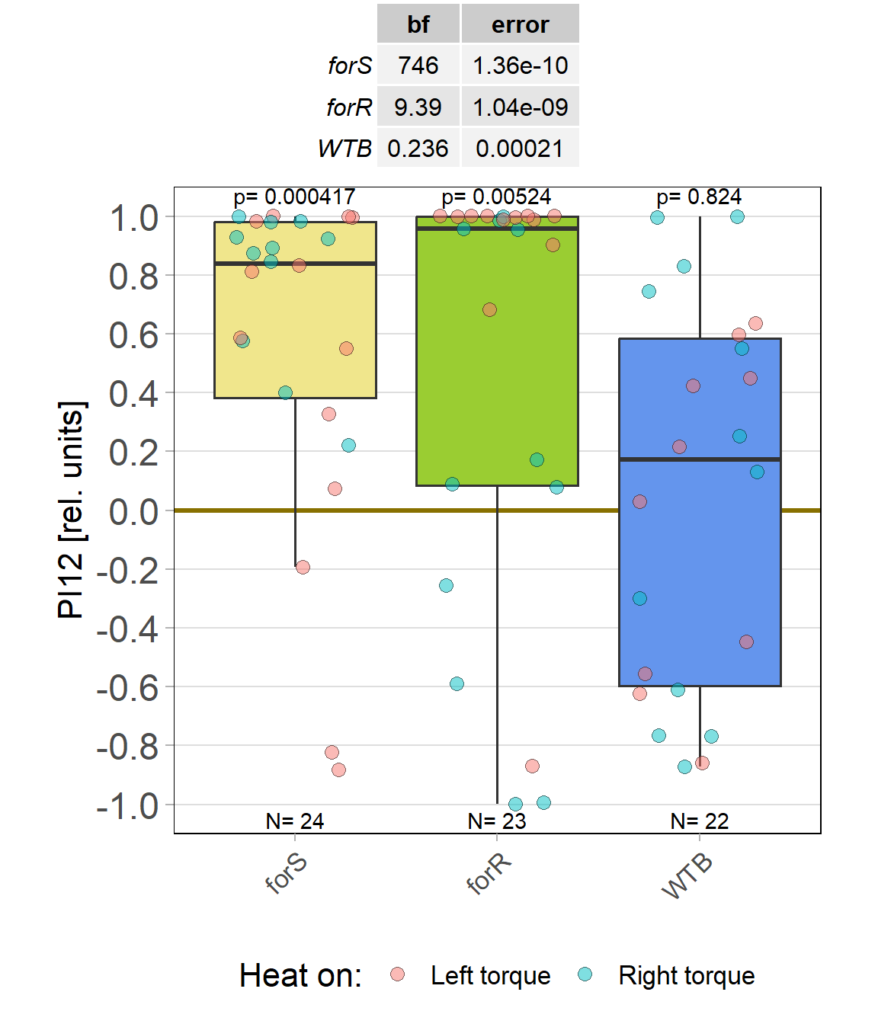
Finally have about half the number of flies needed. It looked like the flies that used the FoxP virgins didn’t fly as well as the other flies, so we dropped that branch and have stopped using them for the crosses. Pooling the FoxP>aPKC/CRISPR flies no increases the N in this group:
Category: Foxp, operant self-learning, PKC | No Comments
Adding flies and fixing figures
on Tuesday, May 21st, 2024 8:56 | by Björn Brembs
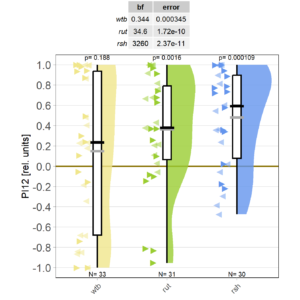
Added more flies to the aPKC knock-out in FoxP neurons. Now the knock-outs are close to zero, but one of the controls, too. Still too early to say much. The figure looks ok now, but the Bayes Factors get chopped off. need to fix this. As I’m, already working on some figures (histograms) in the code, I can fix this as well.

Category: operant self-learning, PKC | No Comments
Finally done: rut and rsh flies better at self-learning
on Monday, January 22nd, 2024 1:36 | by Björn Brembs
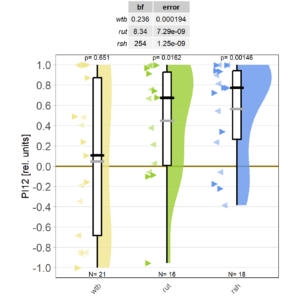
At long last, I got all the flies together that we need for sufficient statistical power. As the preliminary data had indicated, WTB flies don’t learn with the short training, while rut and rsh flies do just fine.
However, this may be due to genetic background effects, so we need to check the CRISPR mutants.
Category: Operant learning, operant self-learning, Radish | No Comments
Starting it back up
on Monday, December 11th, 2023 11:34 | by Björn Brembs
Category: Operant learning, operant self-learning | No Comments
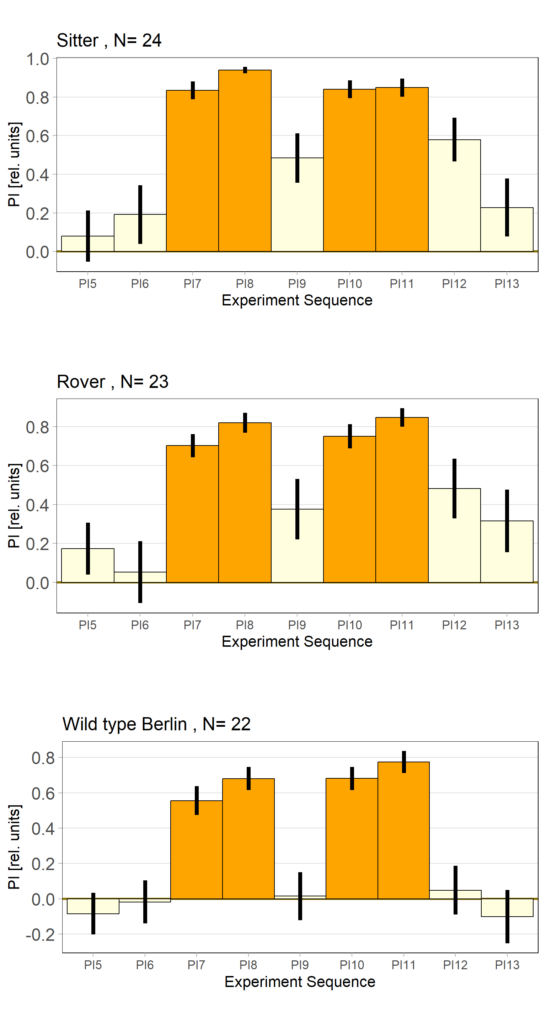
Rover vs. Sitter self learning after 4 minutes training
on Monday, October 16th, 2023 10:39 | by Radostina Lyutova
Category: flight, Habit formation, Memory, Operant learning, operant self-learning, Rover/Sitter | No Comments