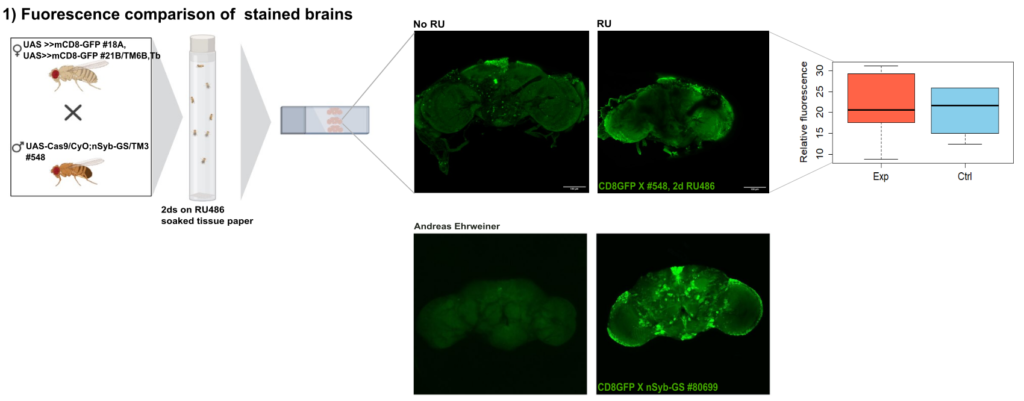
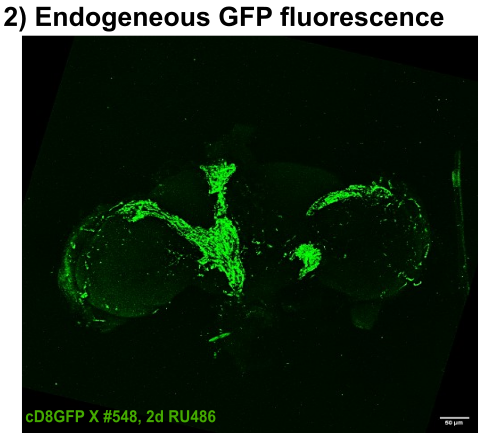
Endogeneous GFP
on Monday, December 22nd, 2025 11:34 | by Julia Schulz
Category: Anatomy, crosses, genetics | No Comments
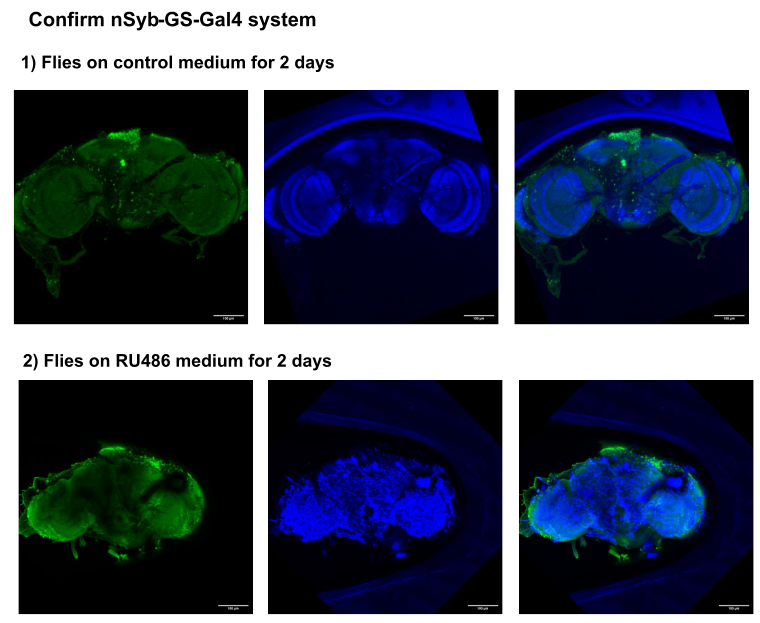
on Monday, December 15th, 2025 1:04 | by Julia Schulz
Category: Anatomy, crosses, genetics | No Comments
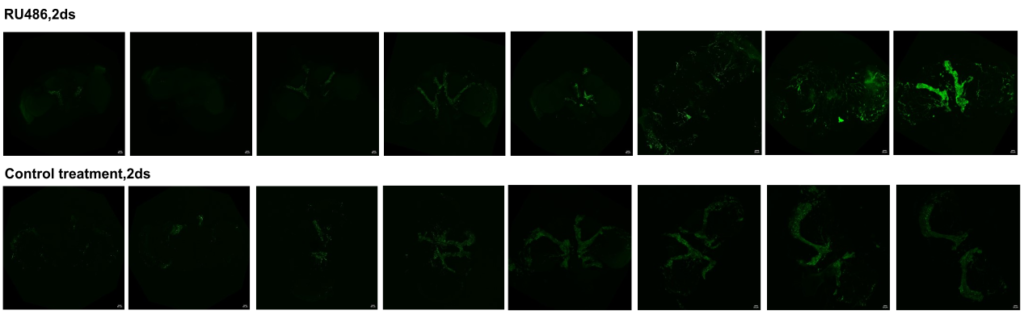
Confocal images
on Monday, December 8th, 2025 12:59 | by Julia Schulz
Category: Anatomy, crosses, Operant learning | No Comments
Confocal images, gain reduced
on Monday, December 1st, 2025 12:42 | by Fridrik Kjartansson
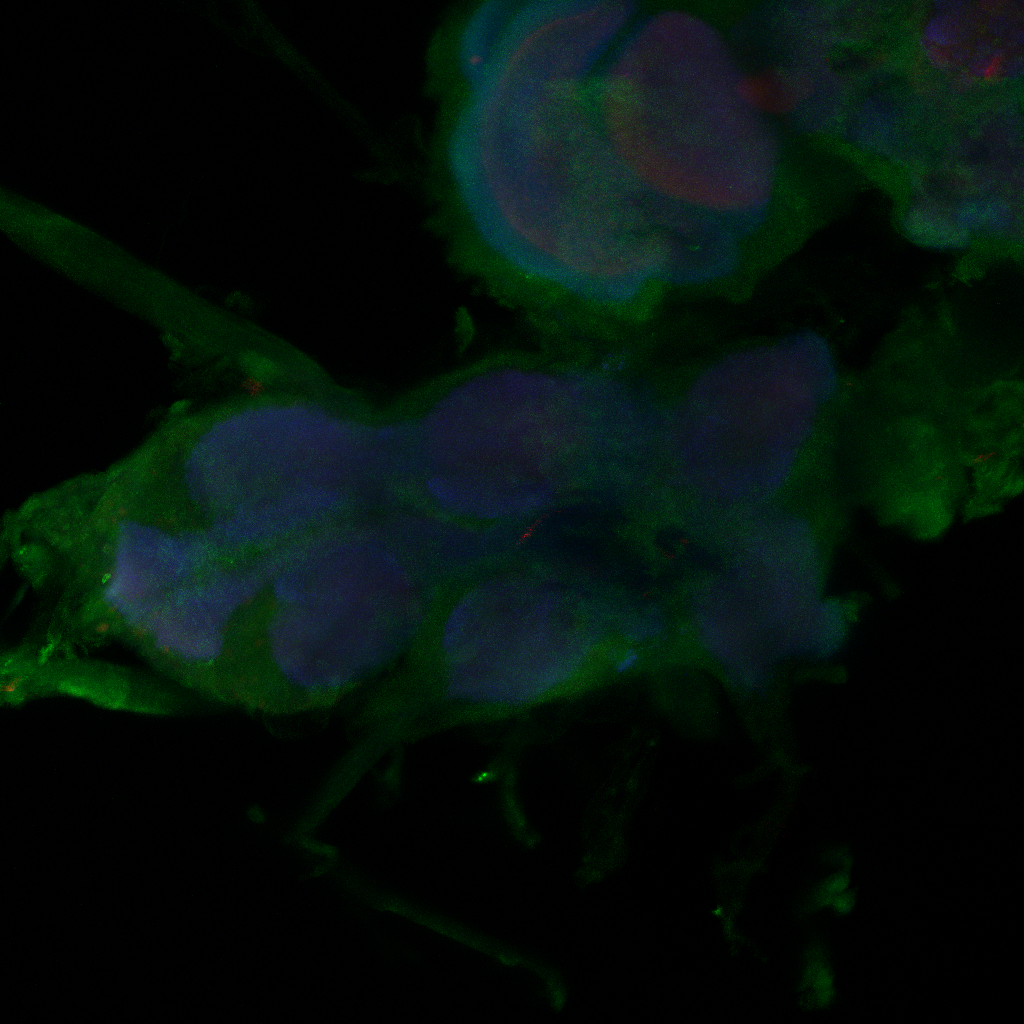
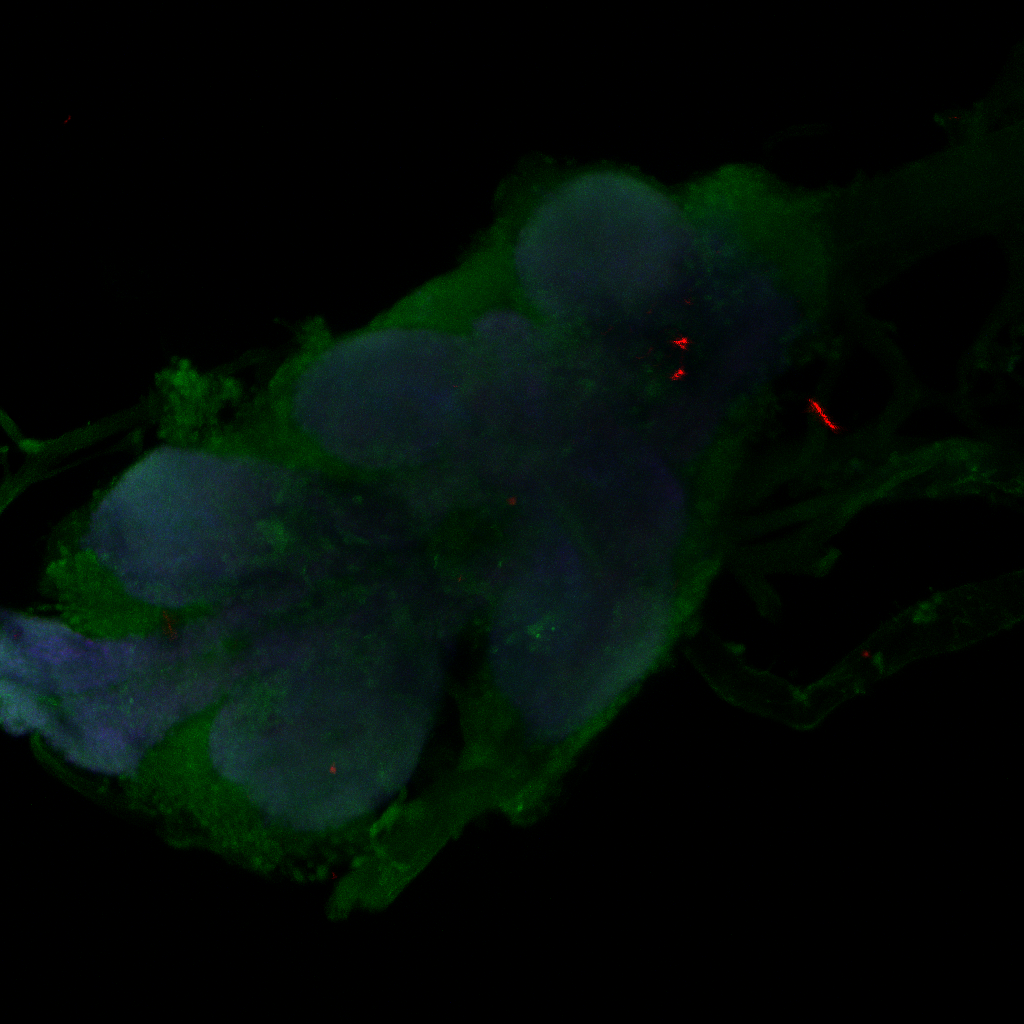
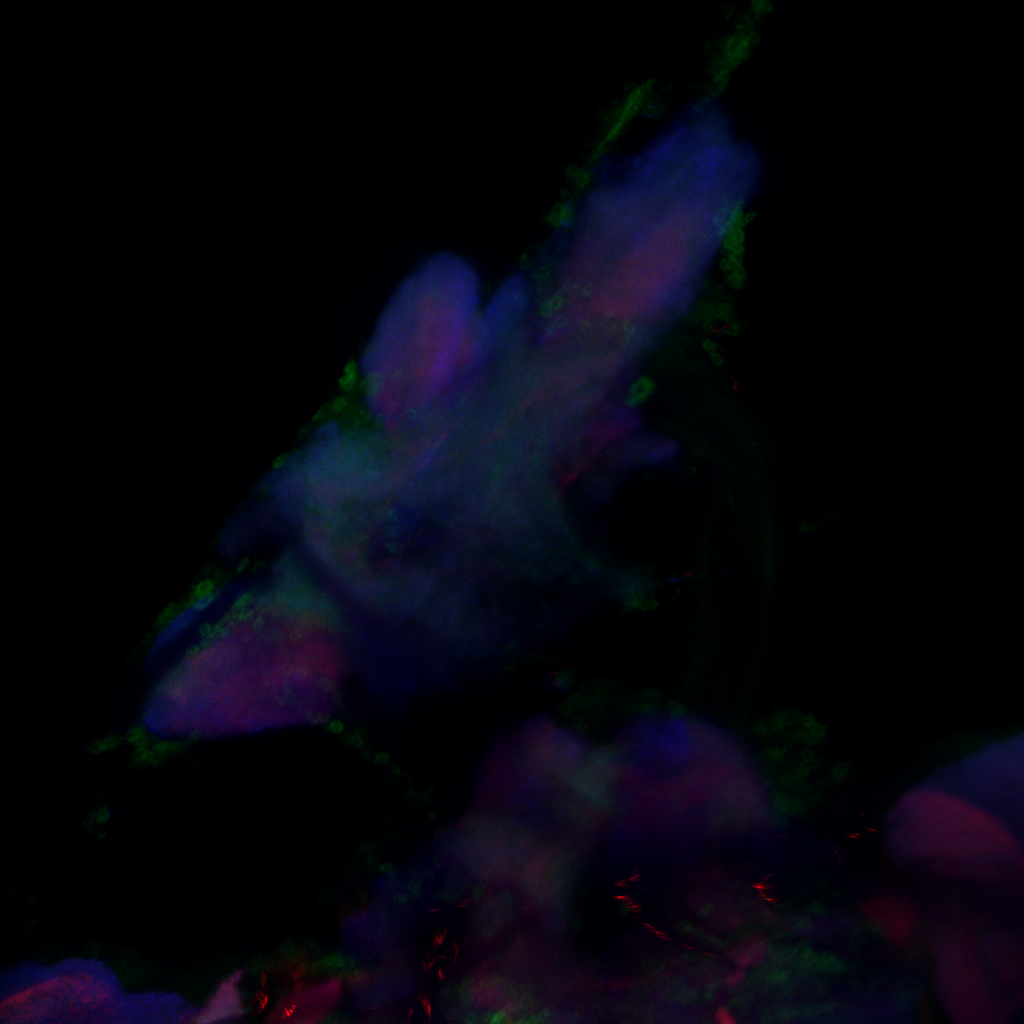
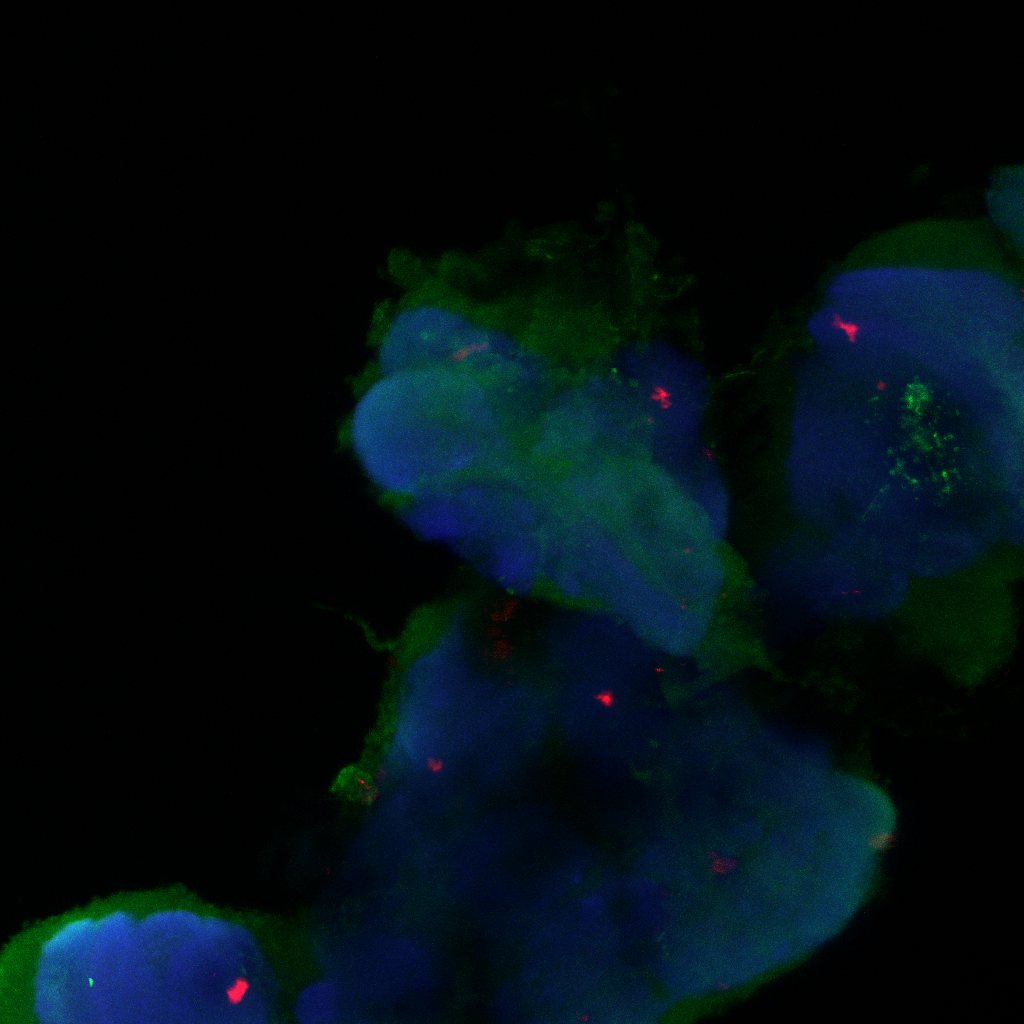
Category: Anatomy, Foxp, genetics, Operant learning, PKC, PKC_localisation | No Comments
Finished Salt (1.5 M) avoidance test in TH-D1-Gal4 larvae under red and blue light (N=20)
on Sunday, November 30th, 2025 9:50 | by Christoph Kumpfmüller

Category: crosses, DAN, genetics, Larve, Mushroom Body | No Comments
Salt (1.5 M) avoidance test in TH-D1-Gal4 larvae under red and blue light [N = 19]
on Sunday, November 23rd, 2025 2:43 | by Christoph Kumpfmüller
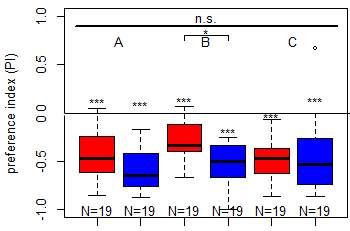
Category: crosses, Larve, Mushroom Body, Optogenetics | No Comments
Finalized Results for the Salt (1.5 M) avoidance tests in DANc1 larvae under red and blue light
on Saturday, November 15th, 2025 2:44 | by Christoph Kumpfmüller
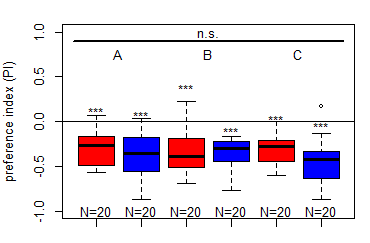
Category: crosses, DAN, Larve, Mushroom Body, Optogenetics | No Comments
Update to the Salt (1.5 M) avoidance tests for the DANc1 larvae tests under red and blue light
on Sunday, November 9th, 2025 5:25 | by Christoph Kumpfmüller
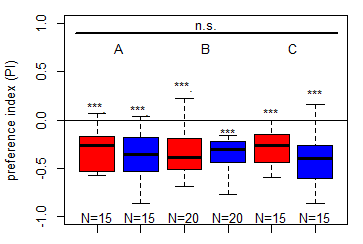
Category: crosses, DAN, Larve, Mushroom Body, Optogenetics | No Comments
First results for Salt (1.5 M) avoidance tests in group A, B and C under red and blue light
on Sunday, November 2nd, 2025 5:27 | by Christoph Kumpfmüller
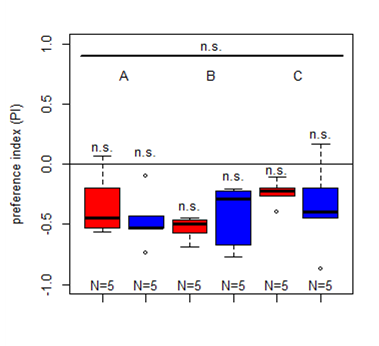
Category: crosses, DAN, Larve, Mushroom Body, Optogenetics | No Comments
Newest crossing scheme for optomotor experiments
on Thursday, August 21st, 2025 12:24 | by Ece Kazgan
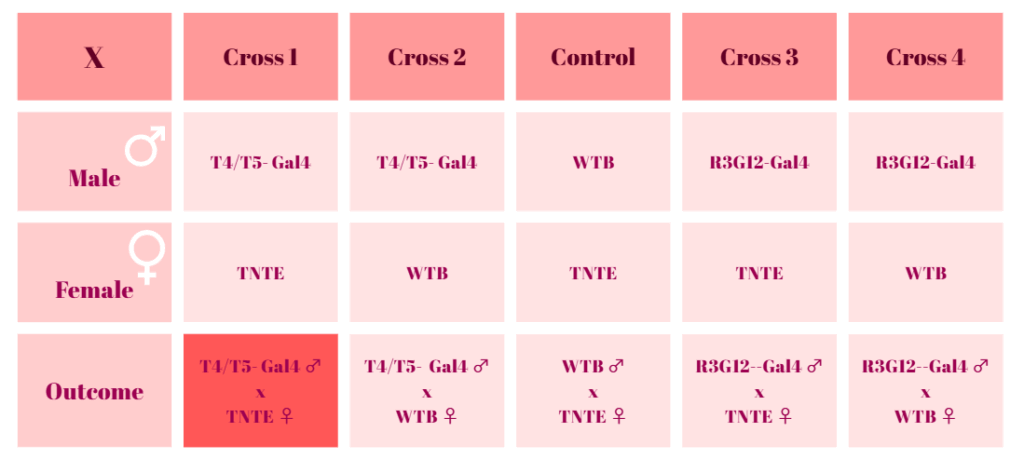
The 1st cross produced eggs, but they did not hatch or become pupae. In all of the 10 vials, eggs were observed, but no pupae. On the other hand, the 2nd cross did not produce any eggs. (The first trial was kept up for 10 days. After 10 days, a new trial began from the same stock. ) Control and the other two crosses produced eggs and hatched new flies.
The same crosses were repeated to ensure the lethality. In the second trial, ‘T4/T5xWTB’ cross produced eggs and pupae. But there is still the possibility that the T4/T5xWTB is also a lethal crossing. ‘T4/T5xTNTE’ cross again produced only eggs but no pupae. The other two crosses produced pupae again and became dark, but haven’t hatched yet. The second attempt is 10 days old.
Category: crosses | No Comments
