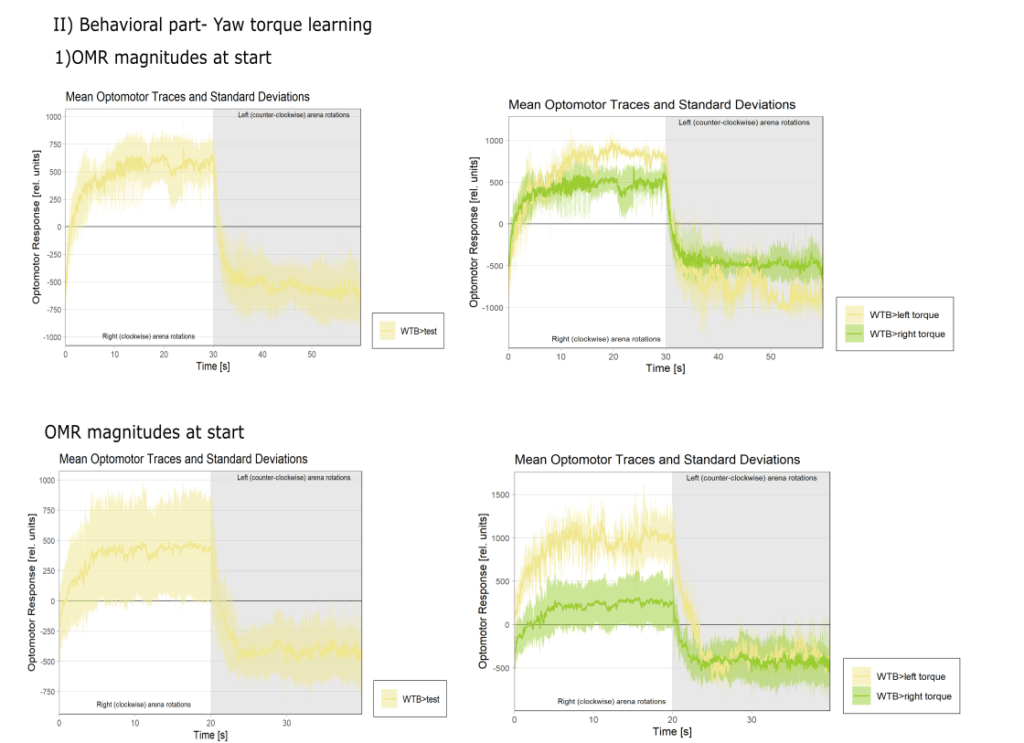
Yaw torque learning
on Monday, September 1st, 2025 12:40 | by Julia Schulz
Category: Operant learning, operant self-learning | No Comments
Molecular work
on Monday, August 4th, 2025 1:59 | by Julia Schulz
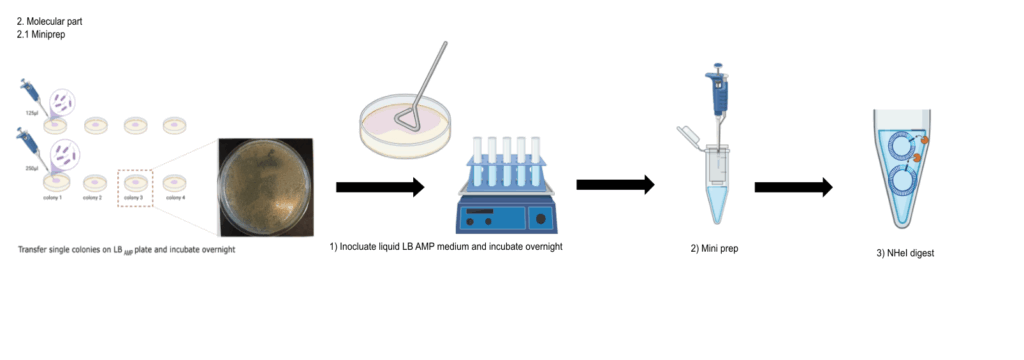
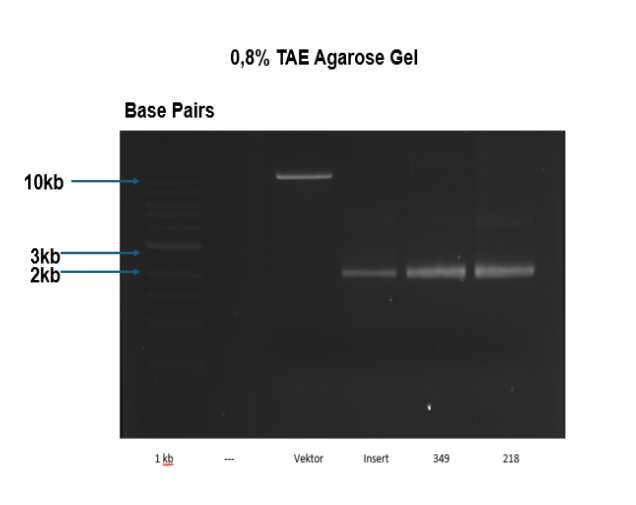
Category: Operant learning, PKC | No Comments
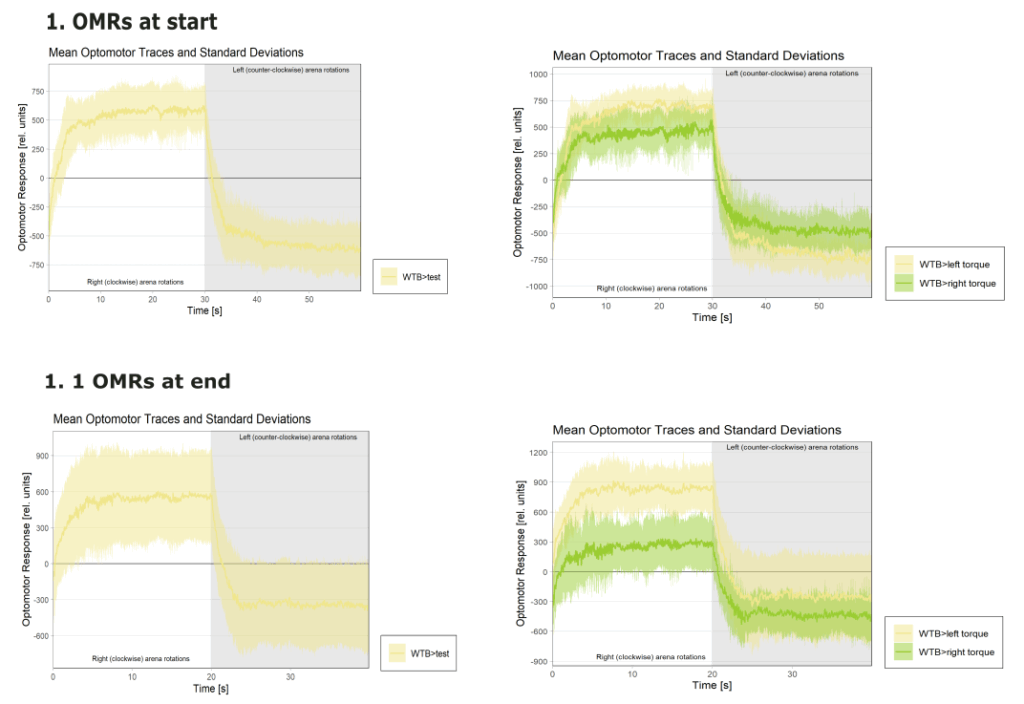
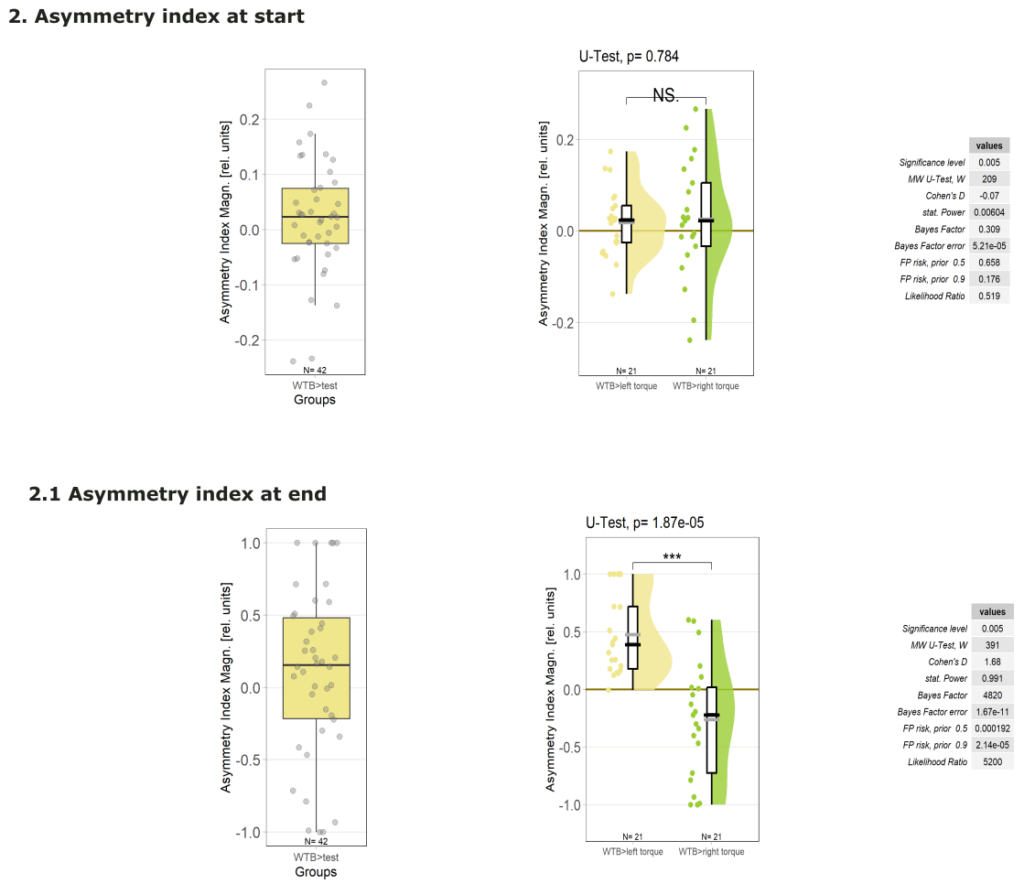
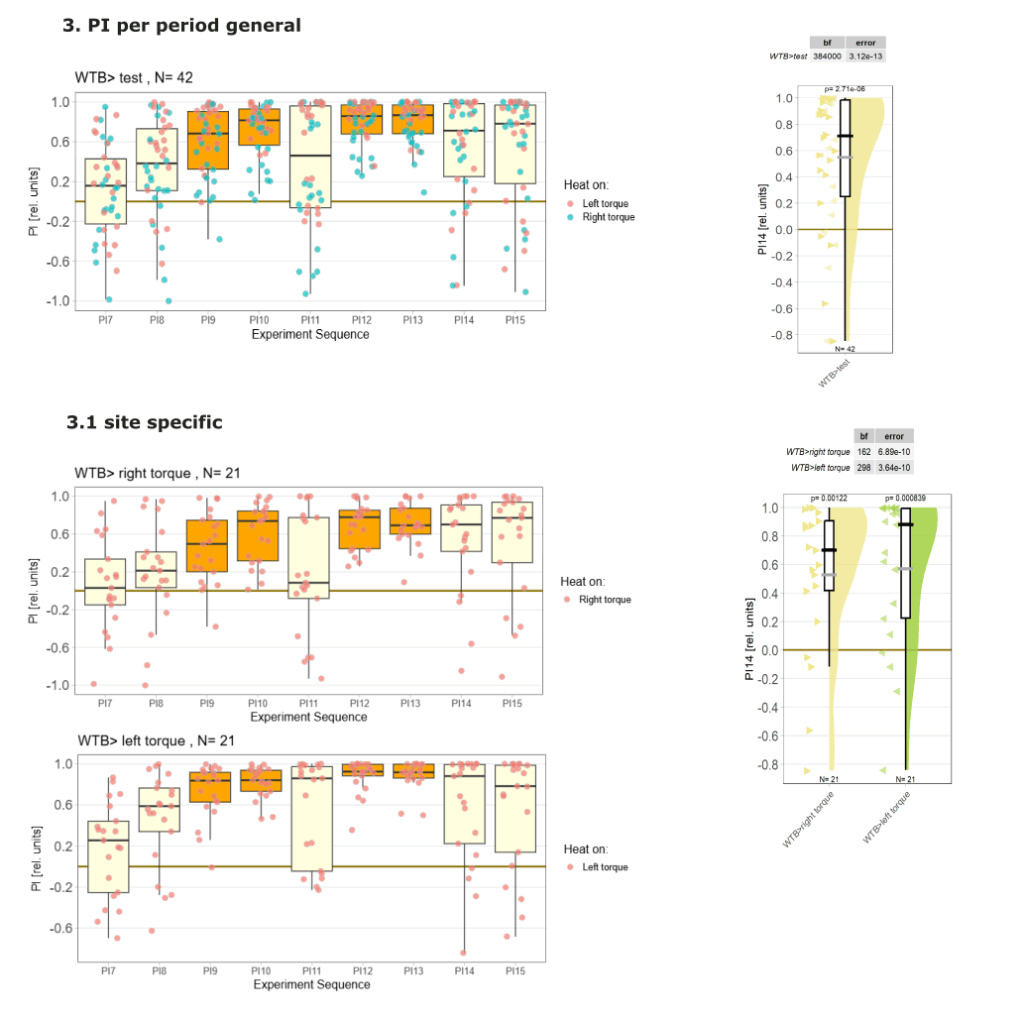
Yaw torque learning
on Monday, August 4th, 2025 1:57 | by Julia Schulz
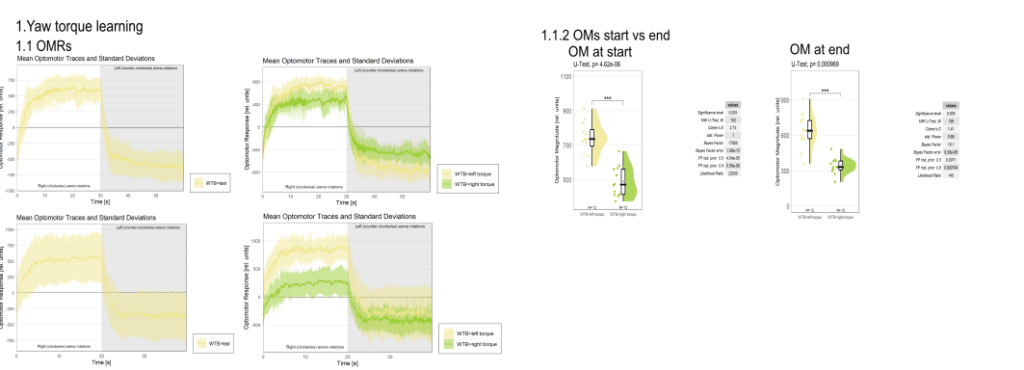
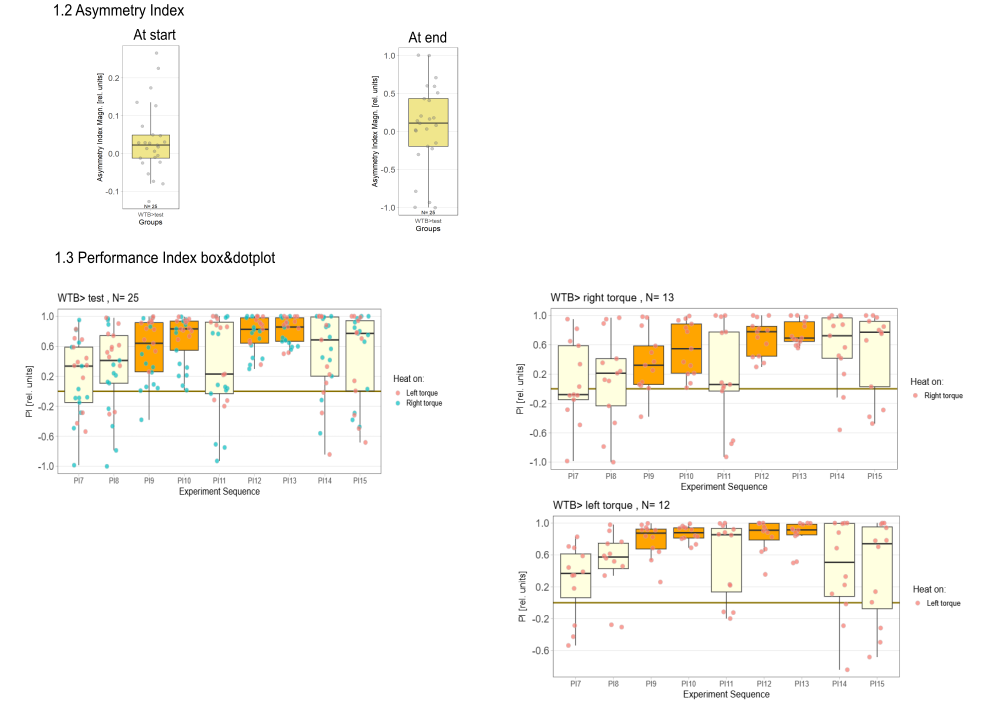
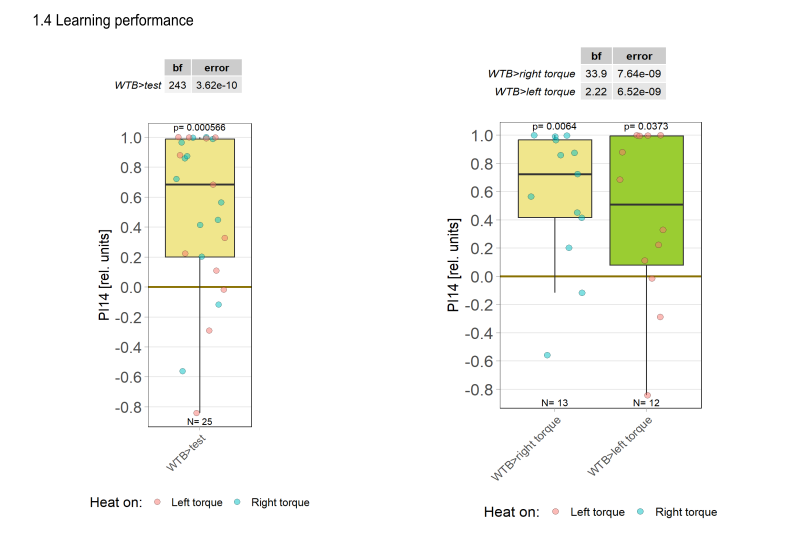
Category: Operant learning, operant self-learning, Optomotor response | No Comments
I) Molecular part
on Monday, July 28th, 2025 1:51 | by Julia Schulz
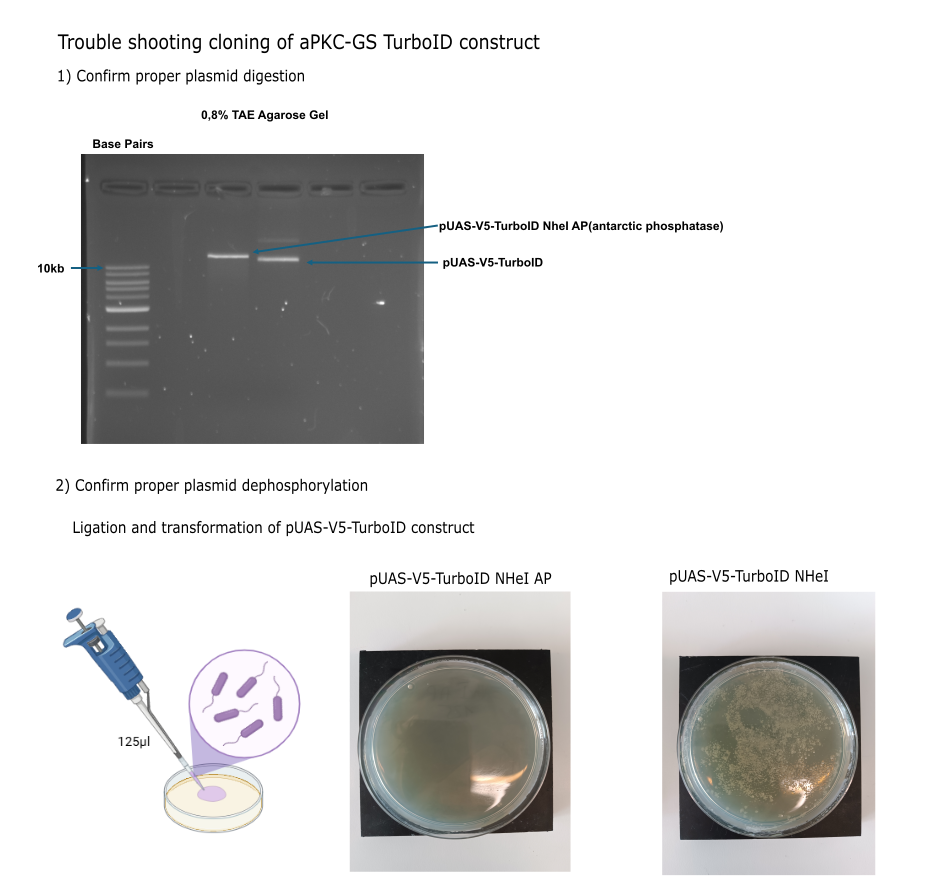
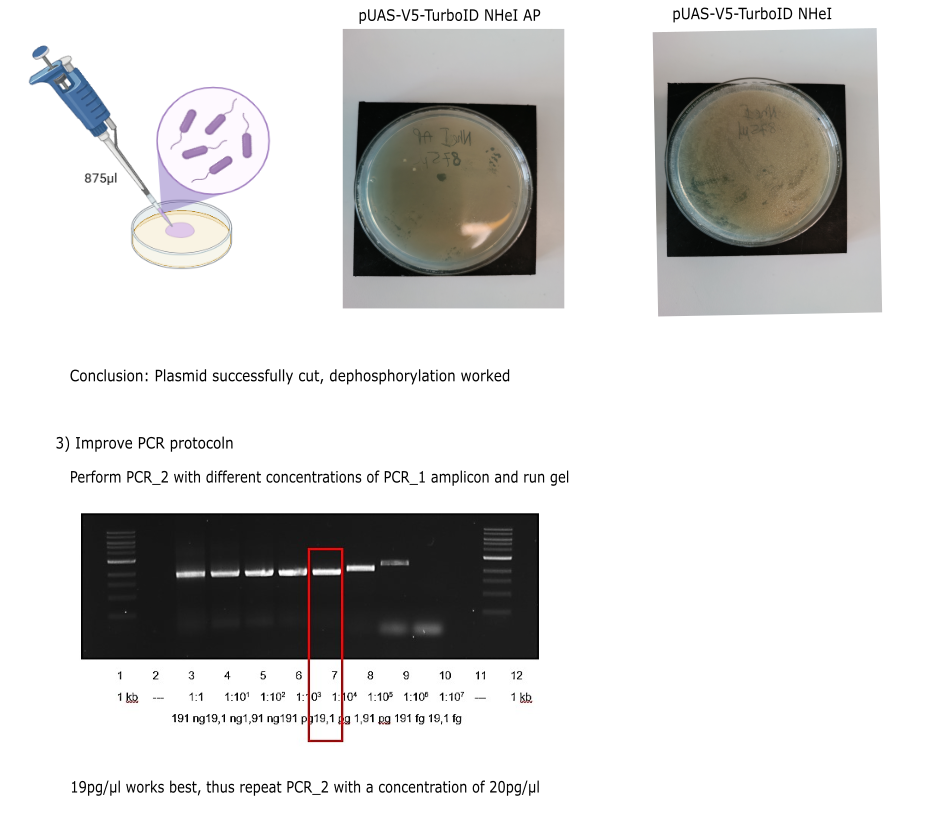

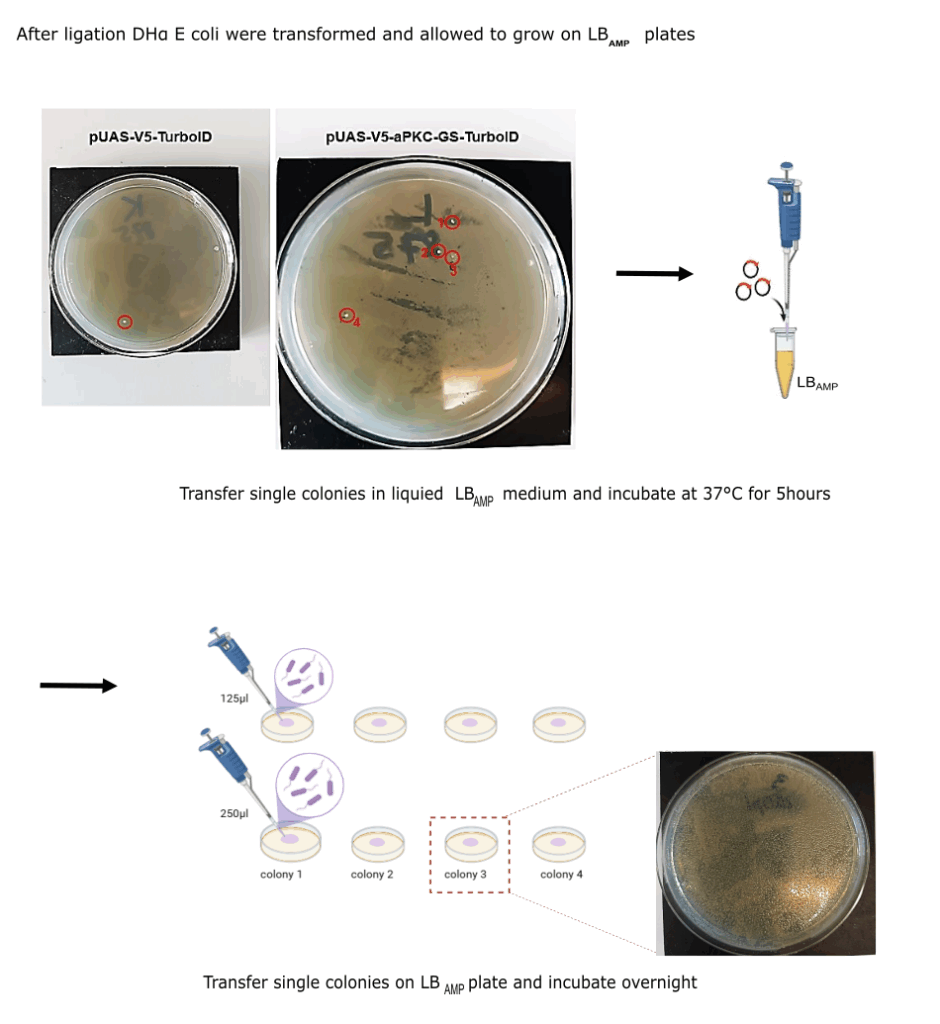
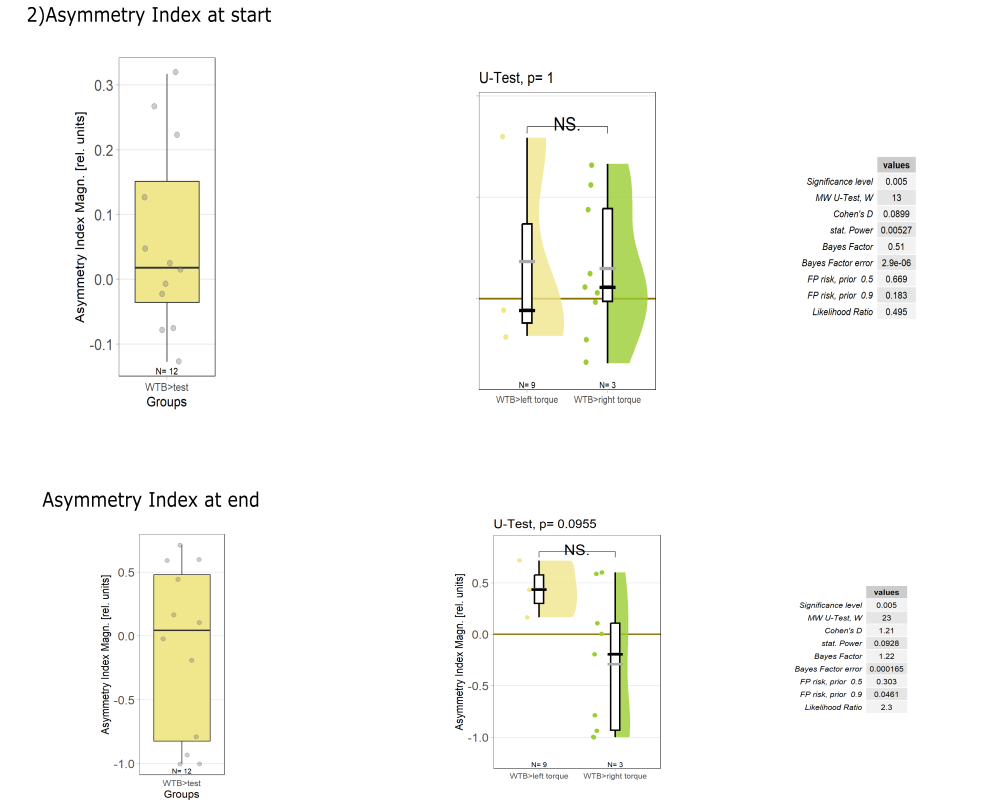
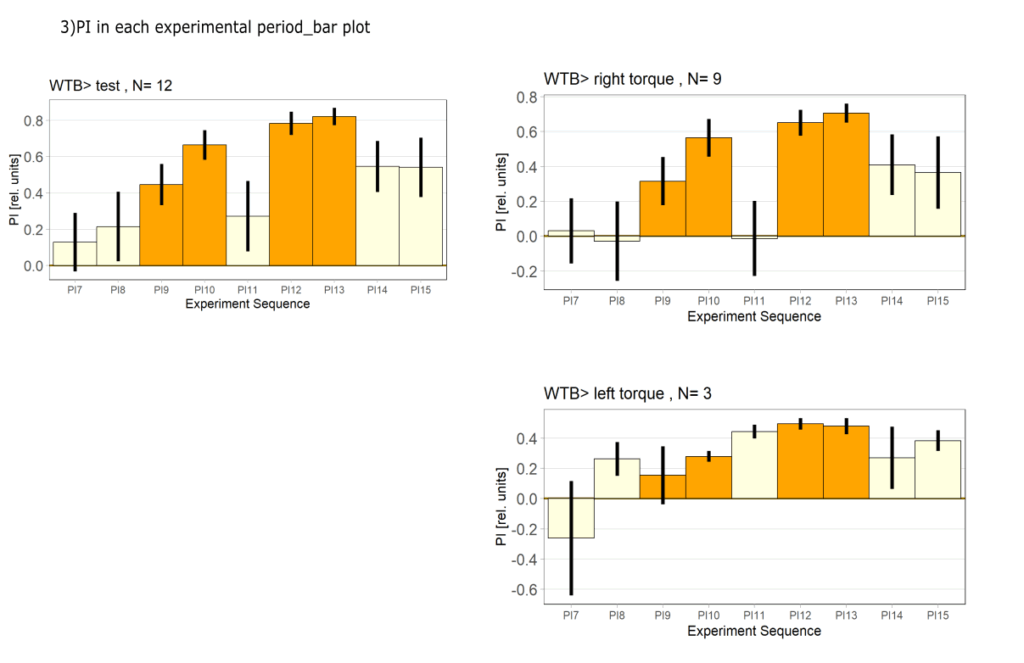
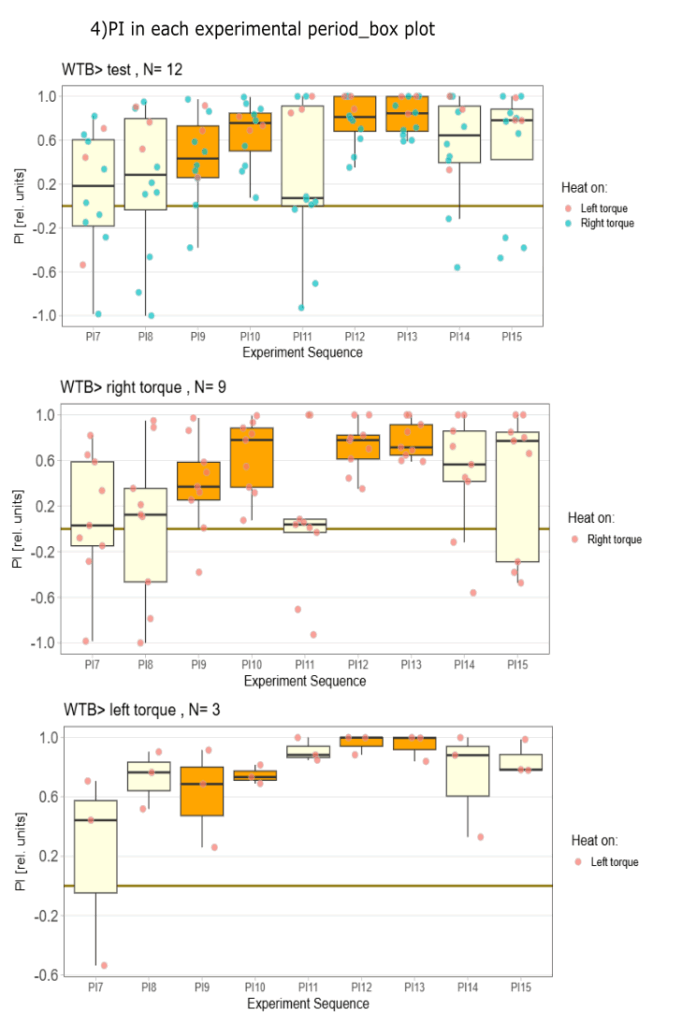
Category: Operant learning, operant self-learning | No Comments
Yt learning_positioning effect
on Monday, June 30th, 2025 11:47 | by Julia Schulz
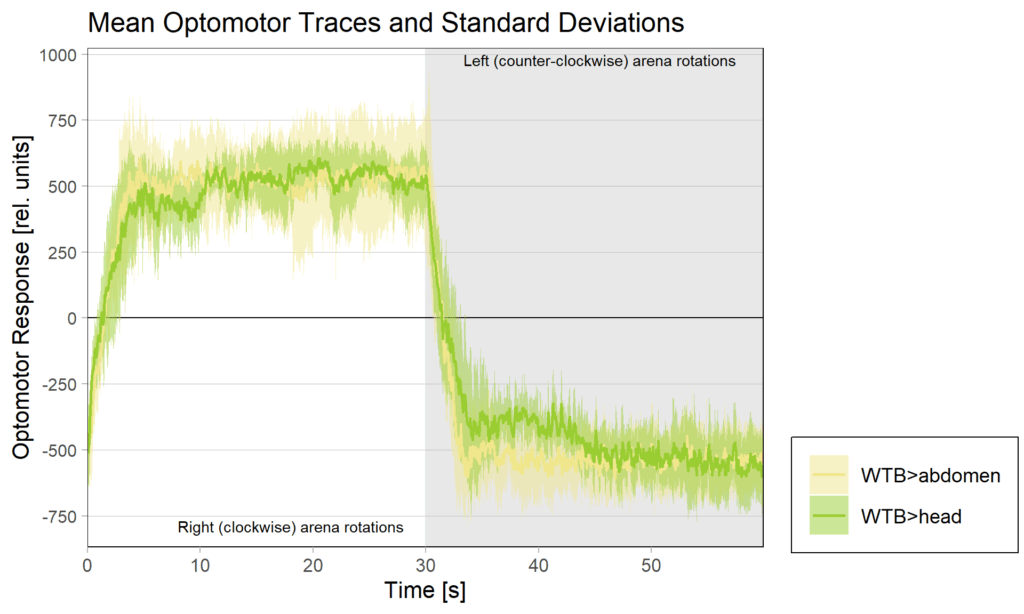
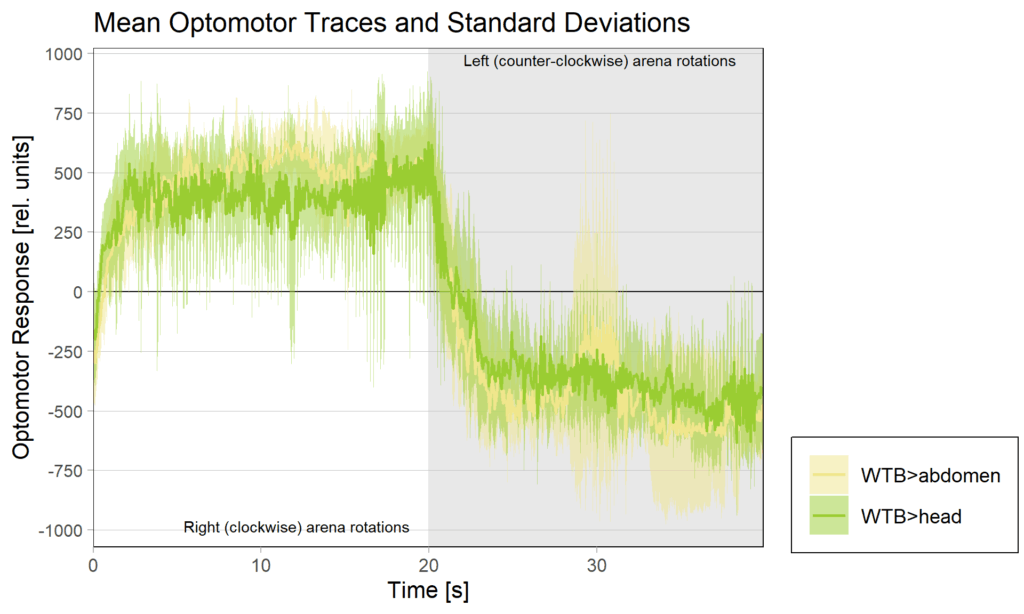
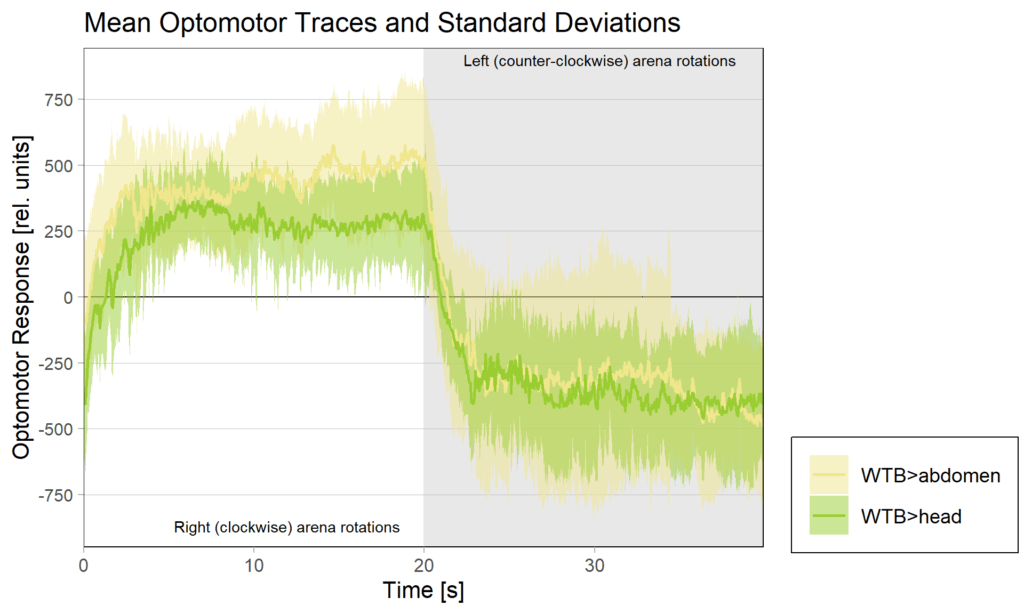
side bias can be compensated for by changing the positioning of the flies within the copper clamp from the medial to the lateral side facing away from the wall.
Category: Operant learning, operant self-learning, Uncategorized | No Comments
YT learning_trouble shooting_body orientation
on Monday, June 23rd, 2025 1:39 | by Julia Schulz
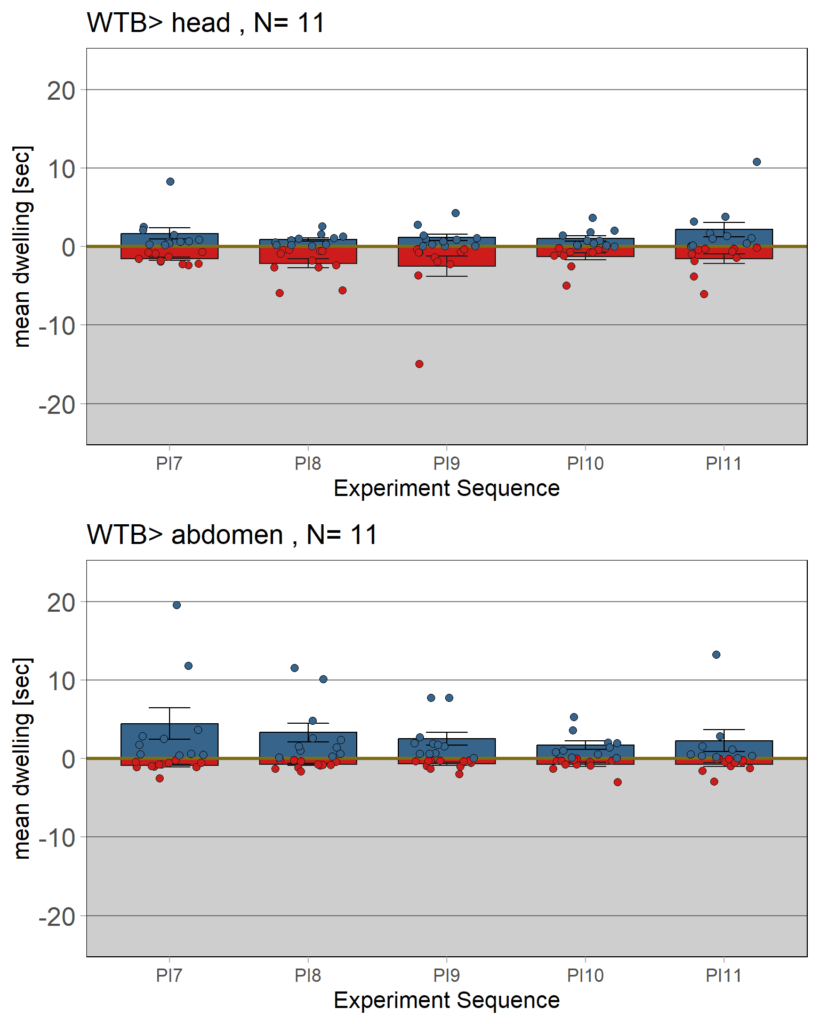
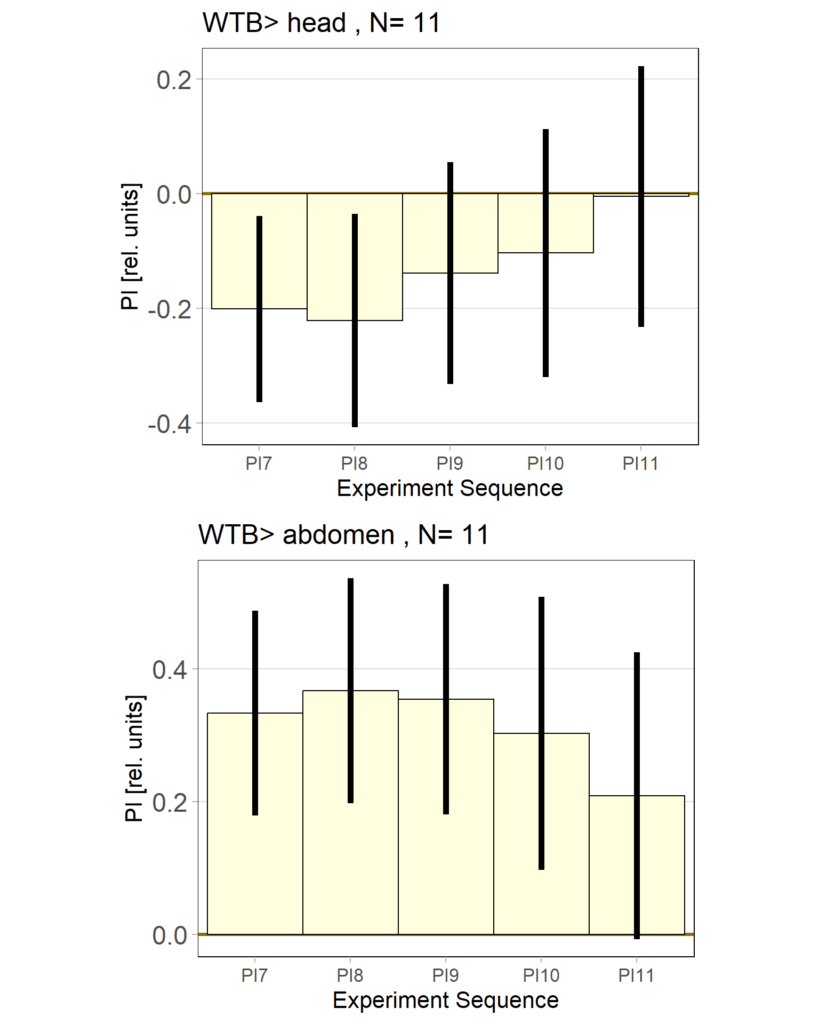
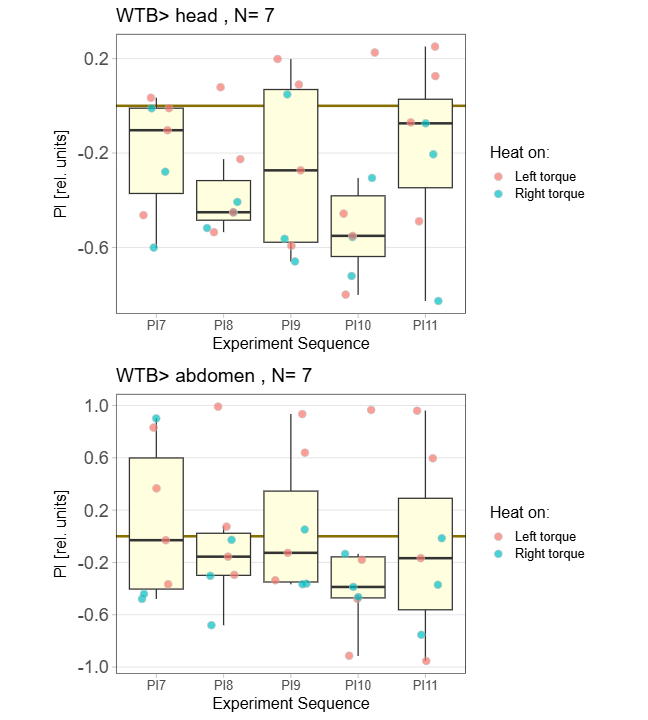
Category: Operant learning, operant self-learning | No Comments
Yt learning_trouble shooting
on Monday, June 23rd, 2025 1:37 | by Julia Schulz
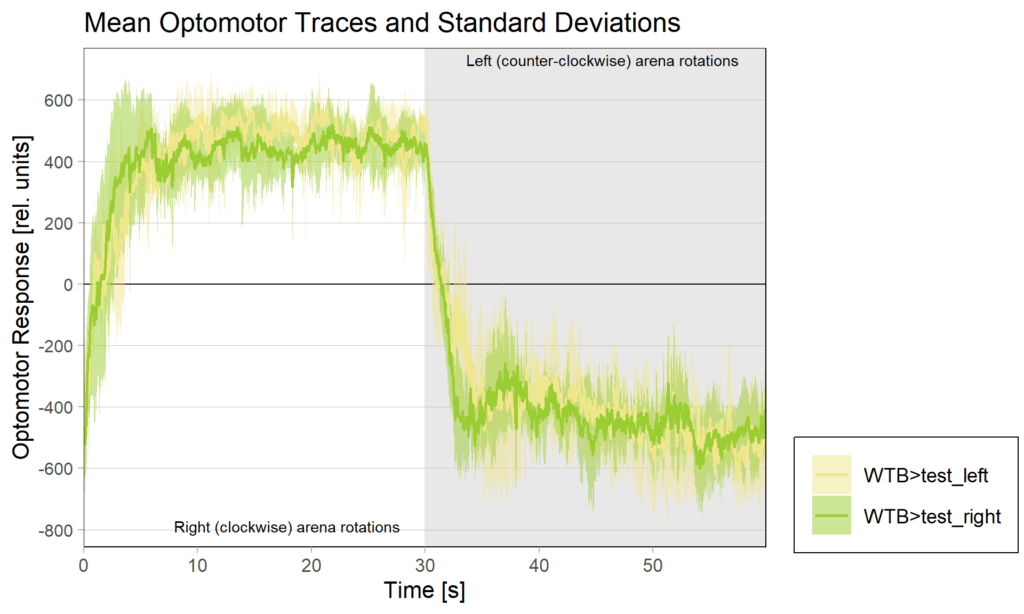
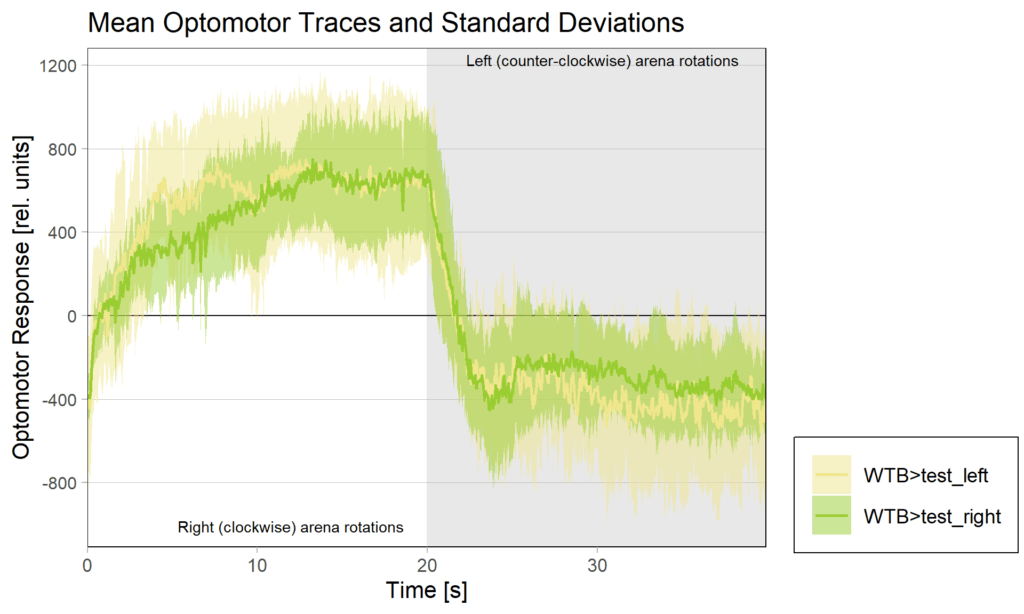
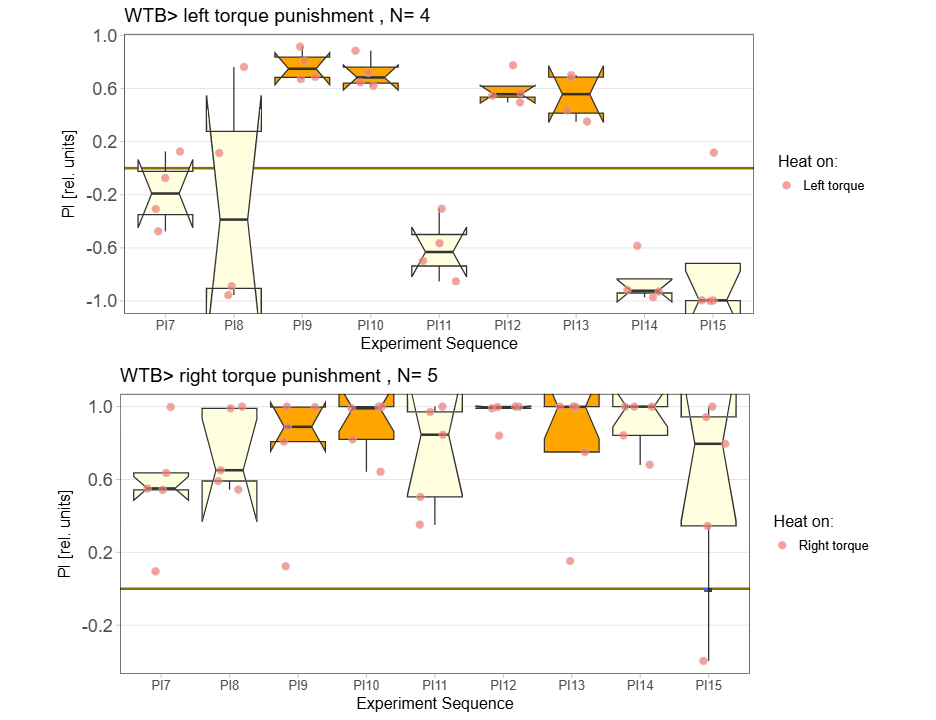
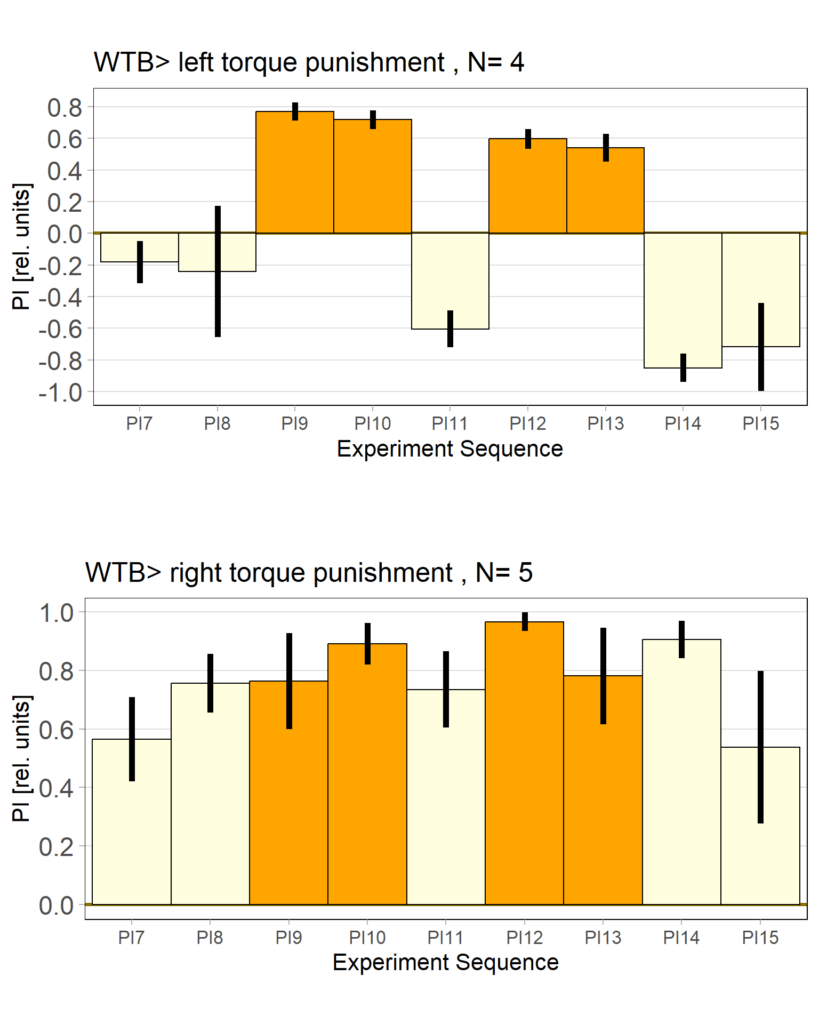
Category: Operant learning, operant self-learning | No Comments
Data sets pooled
on Thursday, May 8th, 2025 8:01 | by Björn Brembs
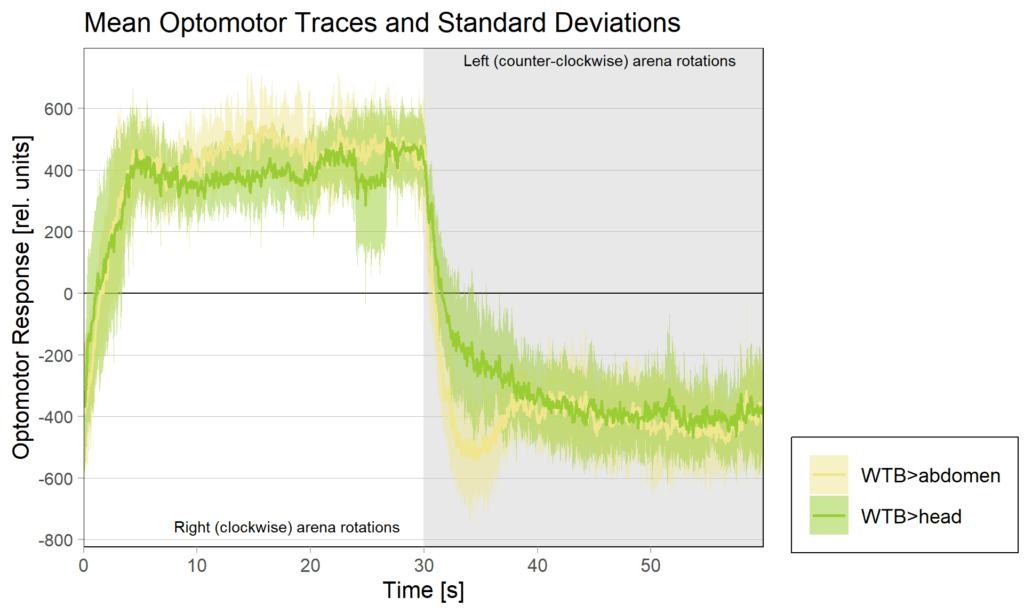
I managed to write some code that shortens Tina’s optomotor periods and then adjusted the metadata between our experiments so that the script would run without any errors. So this is what our combined tests of expressing the foraging RNAi in all neurons looks like:

The full dataset is also available on the UR publication server.
Category: Operant learning, operant self-learning, PKG, Rover/Sitter | No Comments
yaw torque learning_Dtc_40%
on Monday, March 31st, 2025 10:30 | by Julia Schulz
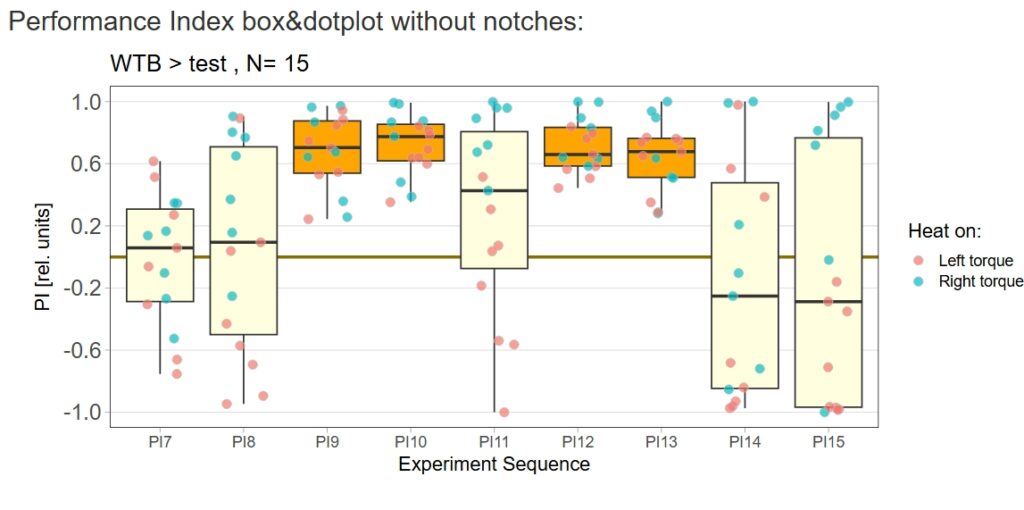
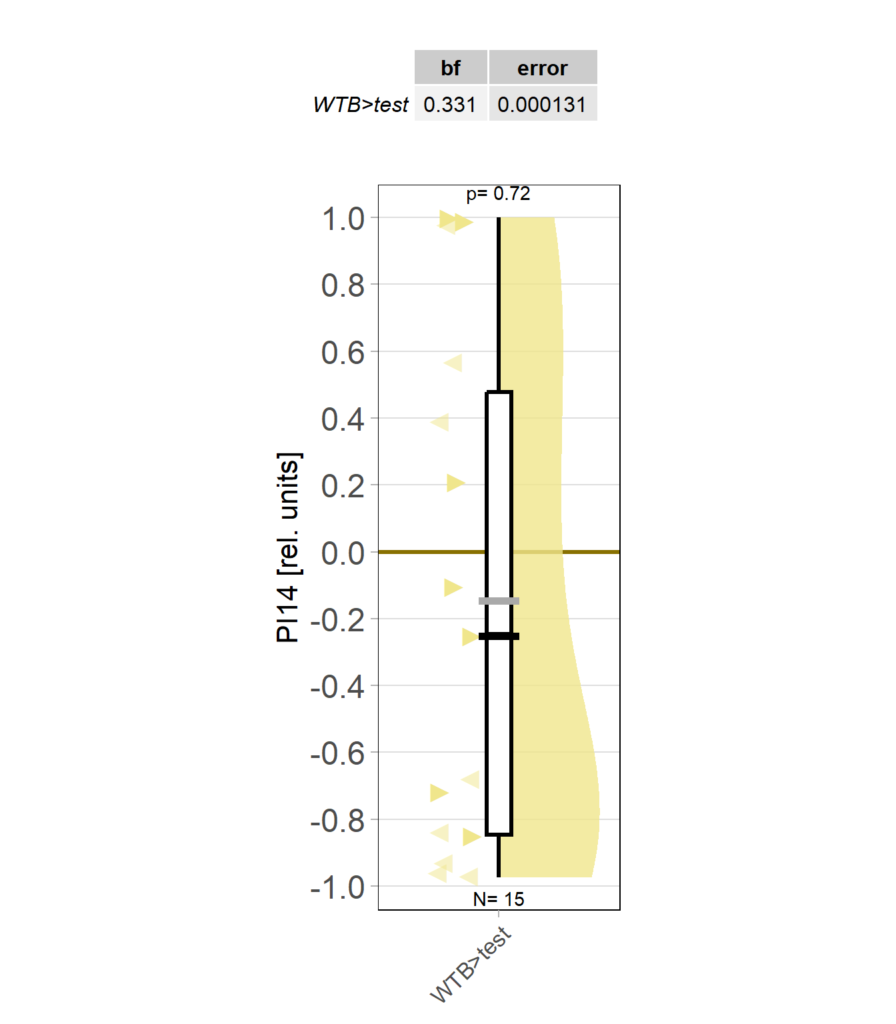
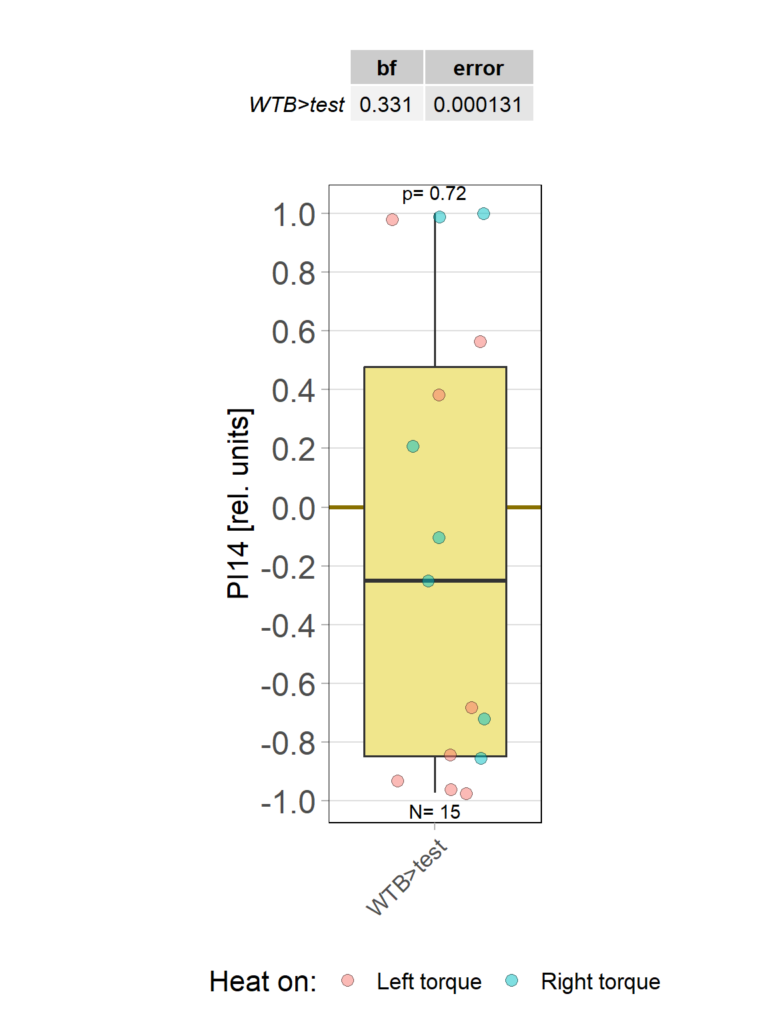
Category: Operant learning, operant self-learning | No Comments
Yaw torque learning_own device
on Monday, March 24th, 2025 11:52 | by Julia Schulz
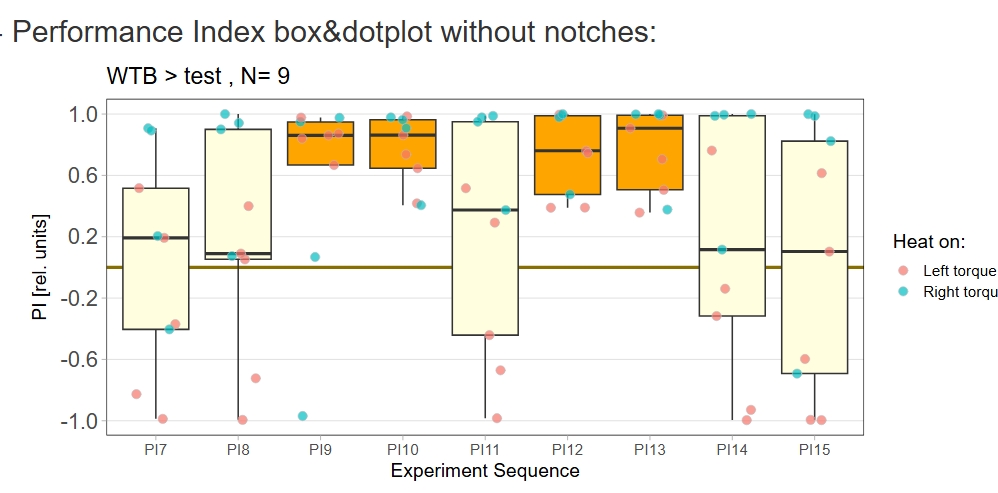
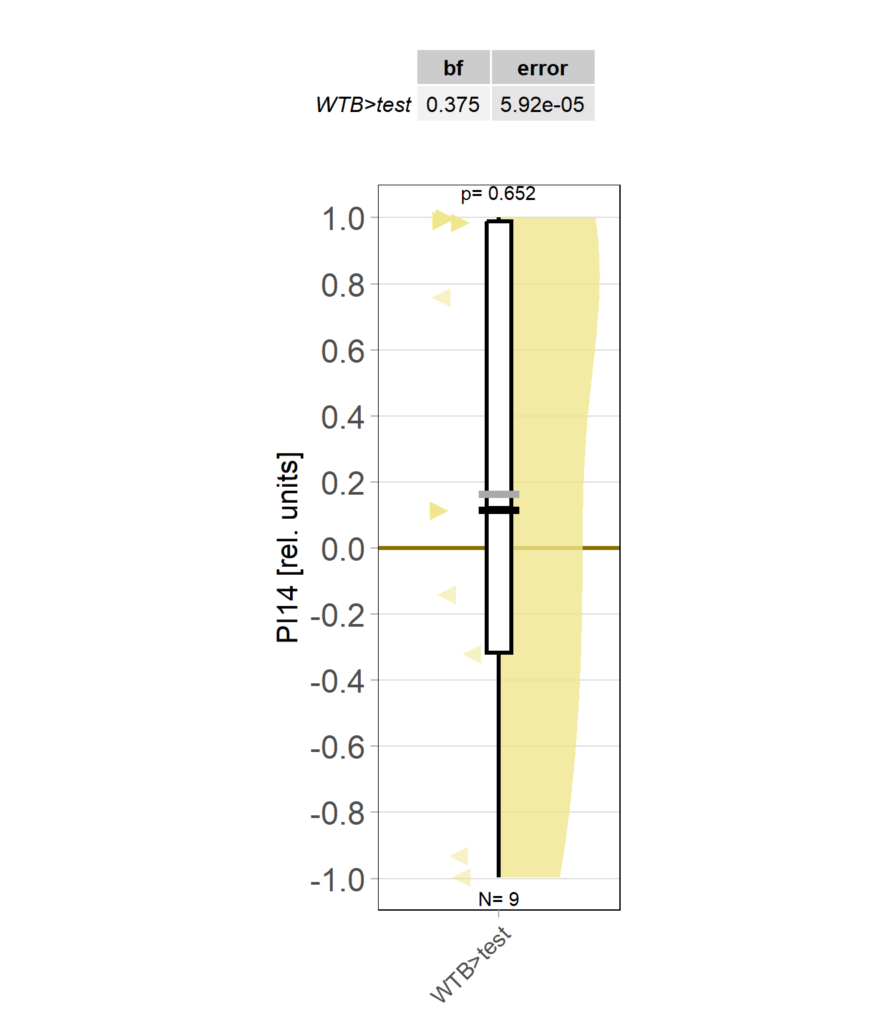
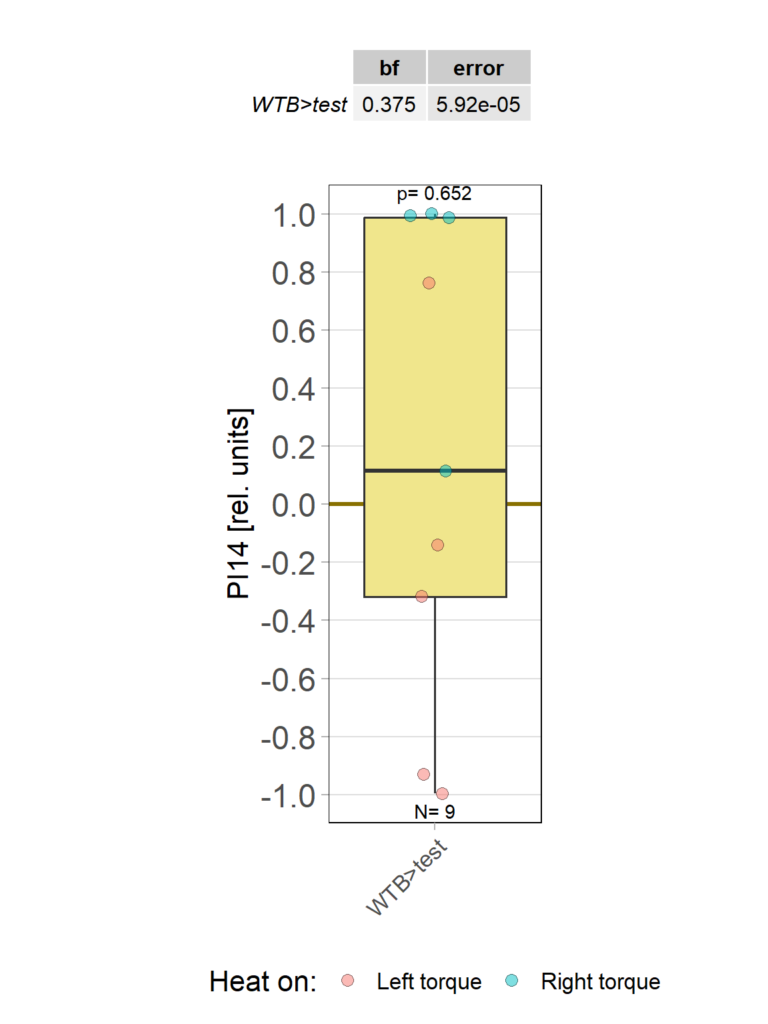
Category: Operant learning, operant self-learning | No Comments
