Salt avoidance under blue and redlight
on Sunday, June 29th, 2025 10:10 | by Eva Schächtl
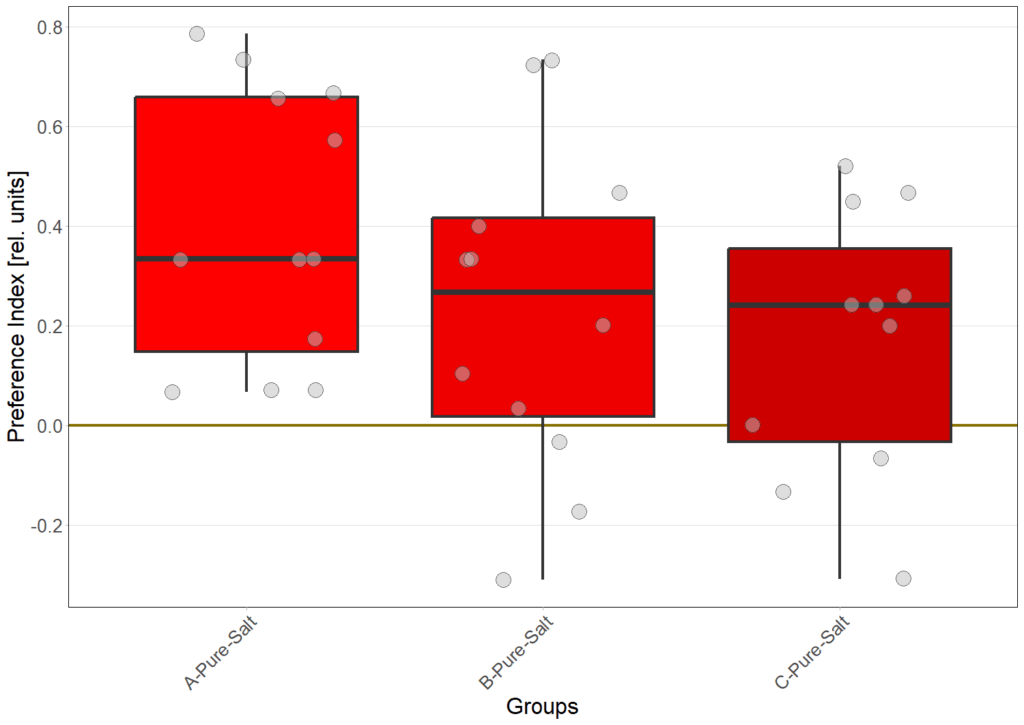
N=12
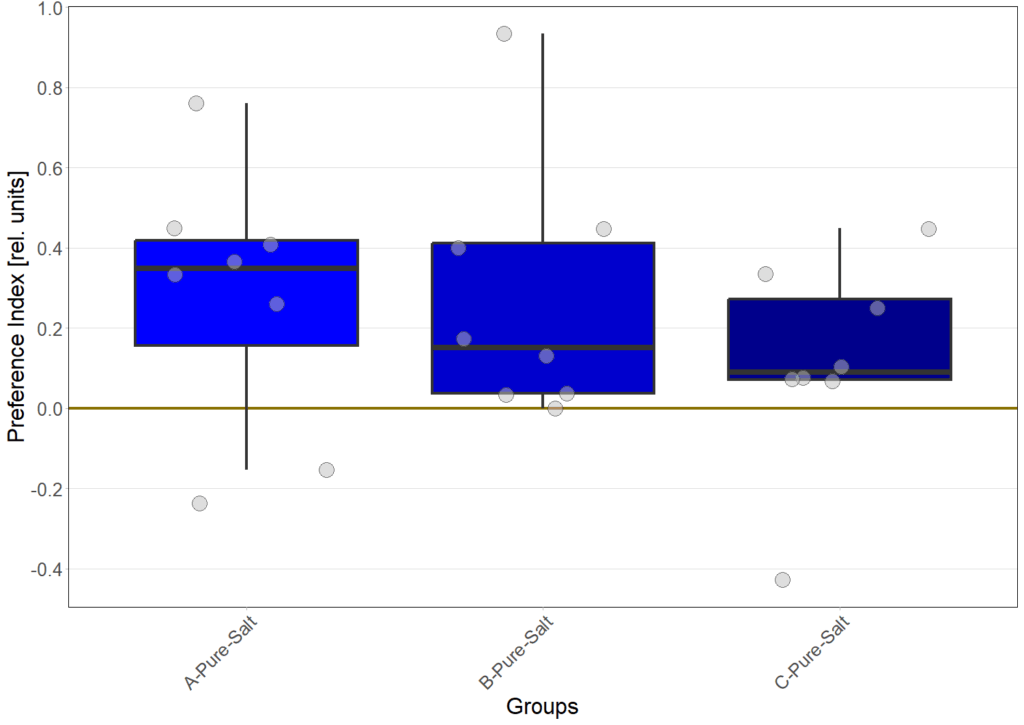
N=8
Category: genetics, Larve, neuronal activation, Optogenetics | No Comments
Gustatory preference results
on Monday, June 2nd, 2025 2:03 | by Eva Schächtl

Gustatory preference under redlight

Gustatory preference under bluelight
Category: Food preference, genetics, Larve, neuronal activation, Optogenetics, Uncategorized | No Comments
Gustatory preference results
on Monday, May 26th, 2025 12:29 | by Eva Schächtl
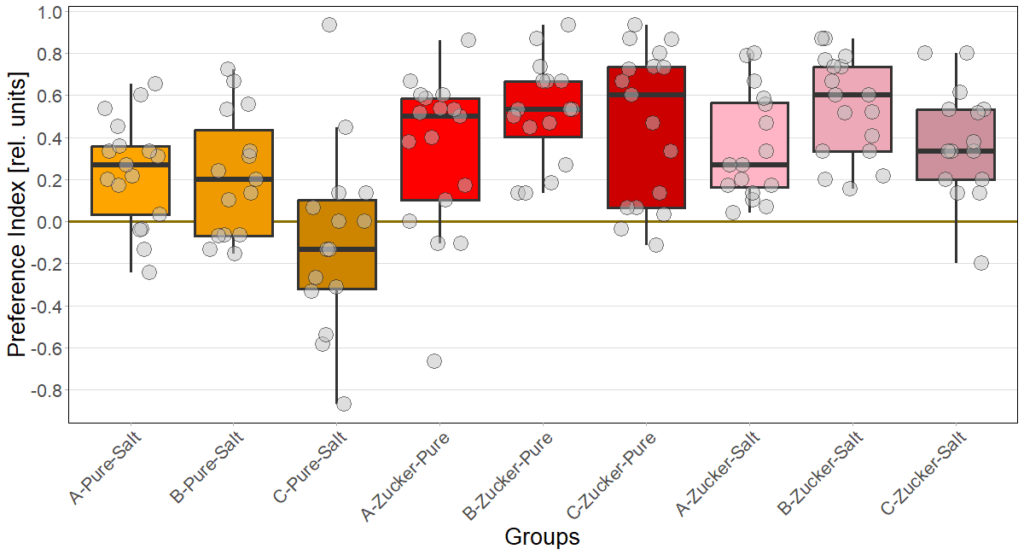
Gustatory preference under redlight
Category: Food preference, genetics, Lab, Larve, neuronal activation, Optogenetics | No Comments
Gustatory preference results
on Sunday, May 18th, 2025 11:45 | by Eva Schächtl
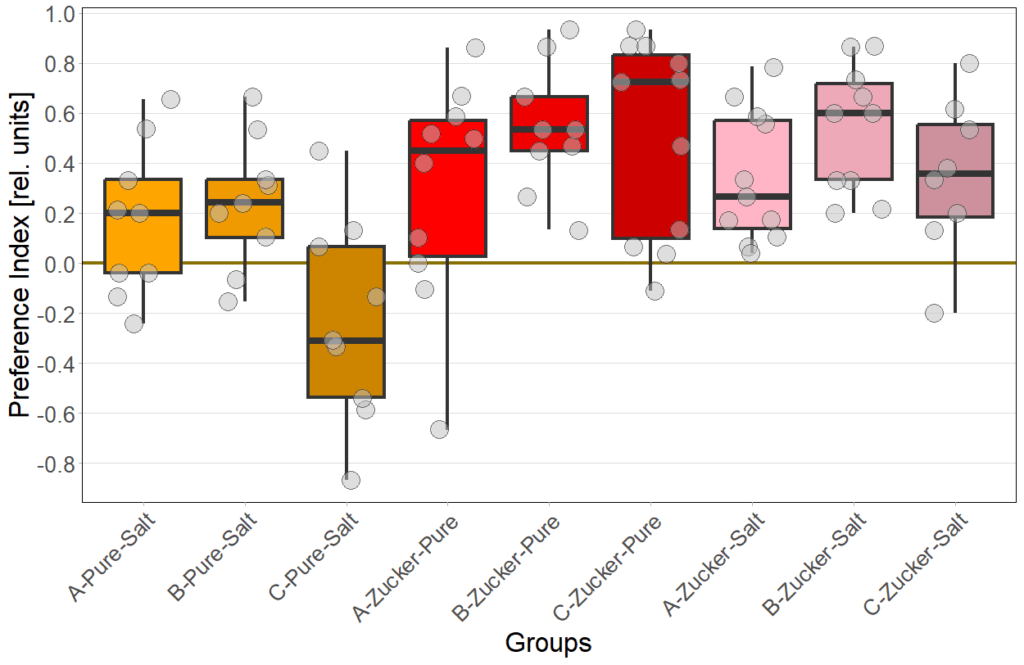
Gustatory preference under redlight
Category: Food preference, genetics, Larve, neuronal activation, Optogenetics, Uncategorized | No Comments
Verification TH-C-AD;TH-D-DBD
on Monday, September 11th, 2023 9:52 | by Luisa Guyton
Category: Anatomy, DAN, genetics, Kenyon cells, Optogenetics | No Comments
Cloning via DNA Assembly
on Friday, September 1st, 2023 7:26 | by Isabel Stark
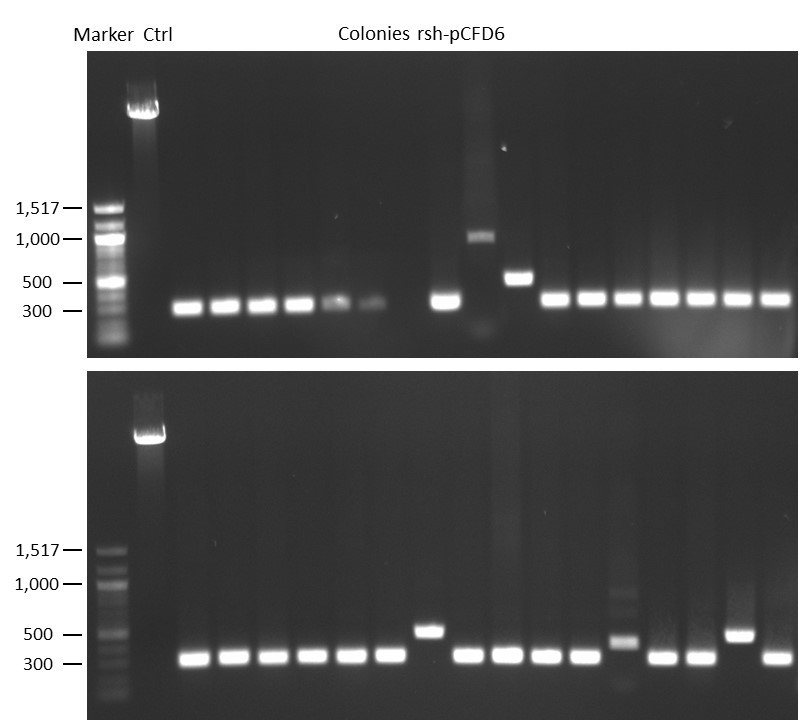
DNA Assembly in a 1:2 ratio of vector to insert with gRNAs of rsh and rut (Q5 and template concentration: 640 pg/µl) and 100ng of pCFD6 BbsI AP (using QuickCIP). Heat-shock (hs) transformation into E. coli (DH5α competent) with 10 µl Assembly Reaction and 100 µl cells.
For Crtl, pCFD6 BbsI AP was wrongly used.
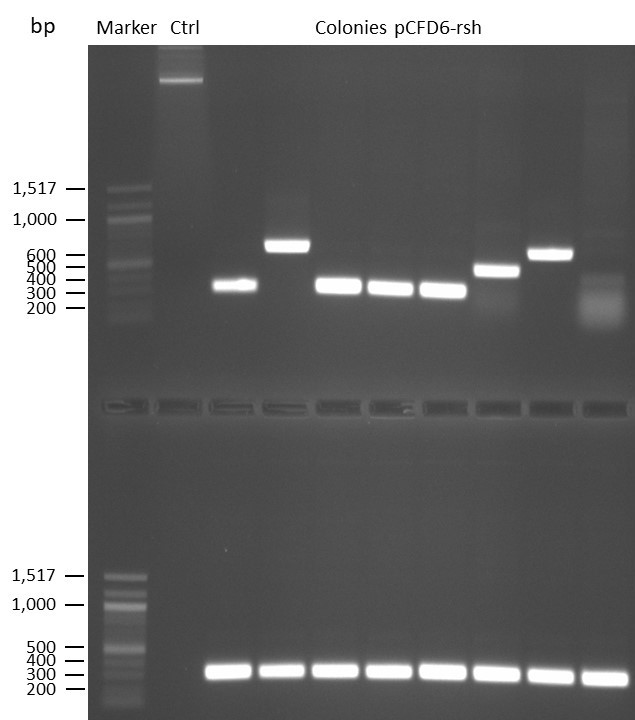
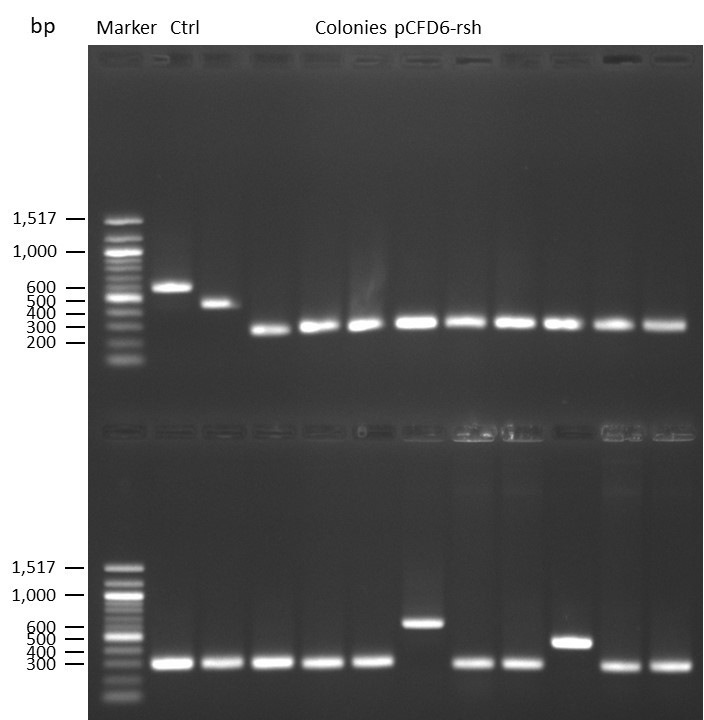
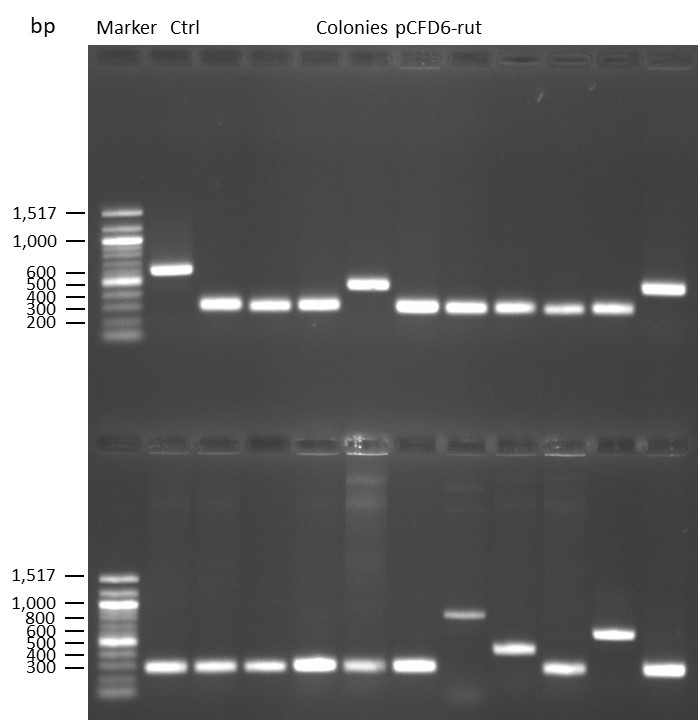
DNA Assembly in a 1:2 ratio of vector to insert with the gRNAs of rsh and rut (Q5 and template concentration: 640 pg/µl) and 100ng of pCFD6 BbsI AP (using FastAP and 2 extraction steps). Heat-shock (hs) transformation into E. coli (DH5α competent) with 10 µl Assembly Reaction and 100 µl cells.
For Crtl, pCFD6 BbsI AP was used in reaction.
Category: genetics, Memory, Operant learning, operant self-learning, Radish | No Comments
Line verification SS56699
on Tuesday, August 29th, 2023 7:50 | by Luisa Guyton
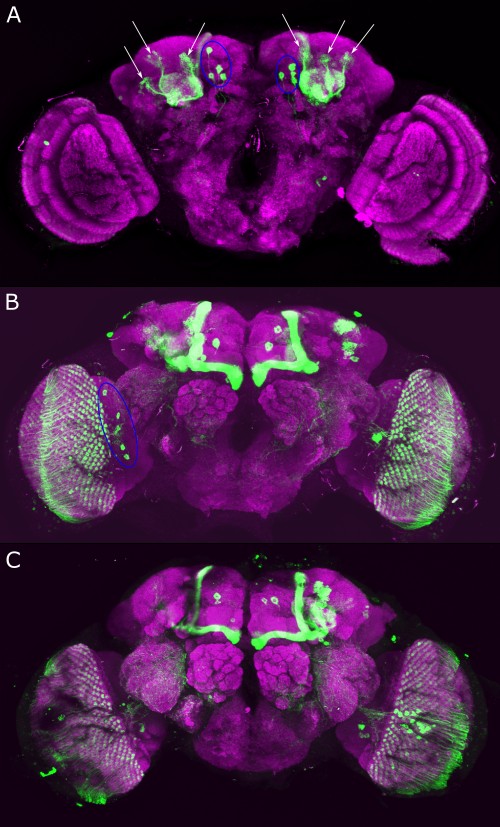
After dissecting brains from the GAL4 driver line SS56699 with GFP staining and finding no fluorescence, I dissected them again and the immunohistochemical staining showed fluorescence. To check that the correct neurons were stained, I compared the images with the image in the paper by Hulse et al (2021; https://doi.org/10.7554/eLife.66039) . The three PPL1 dopaminergic dorsal fan-shaped body tangential neurons per hemisphere are stained, but there are some additional unidentified neurons (presumably PPM1 neruons) visible.
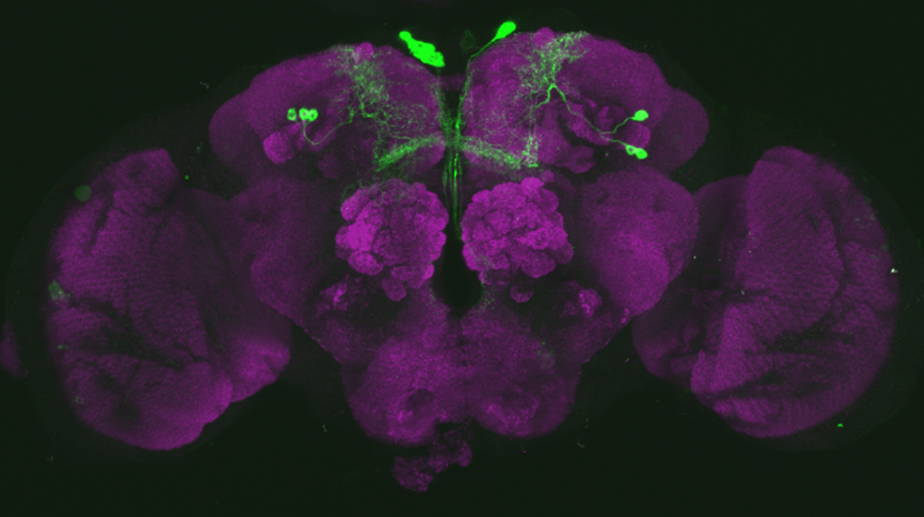
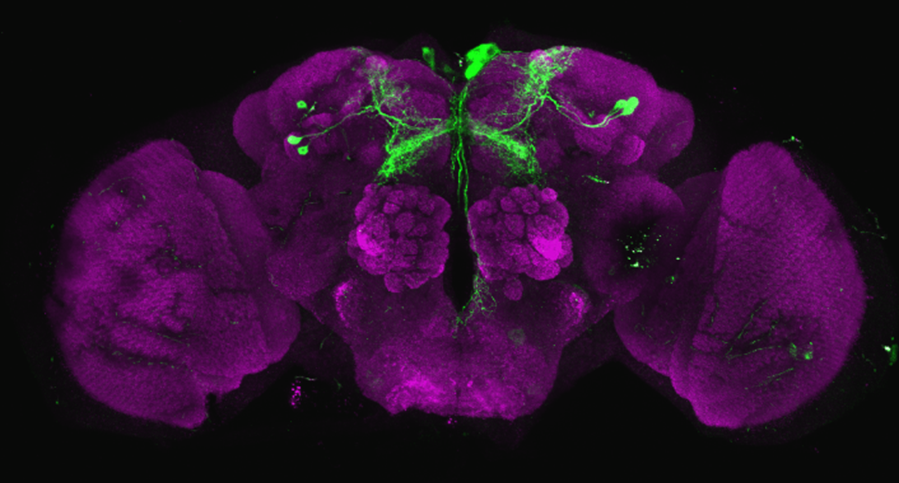
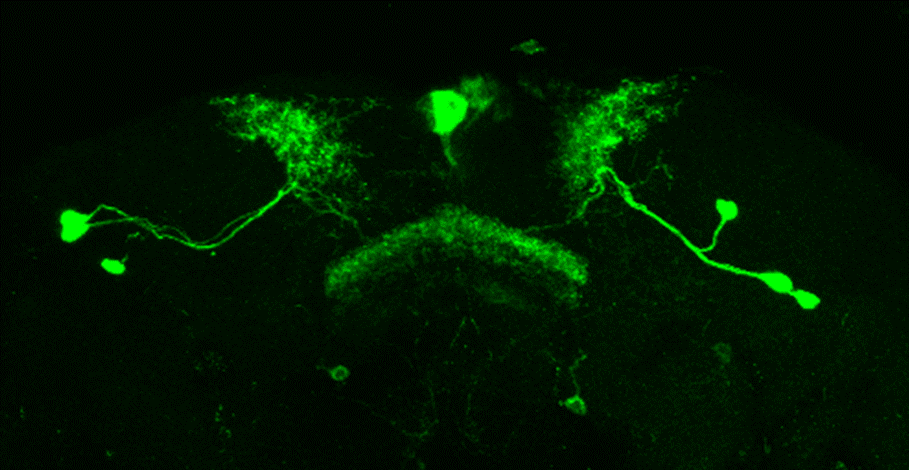
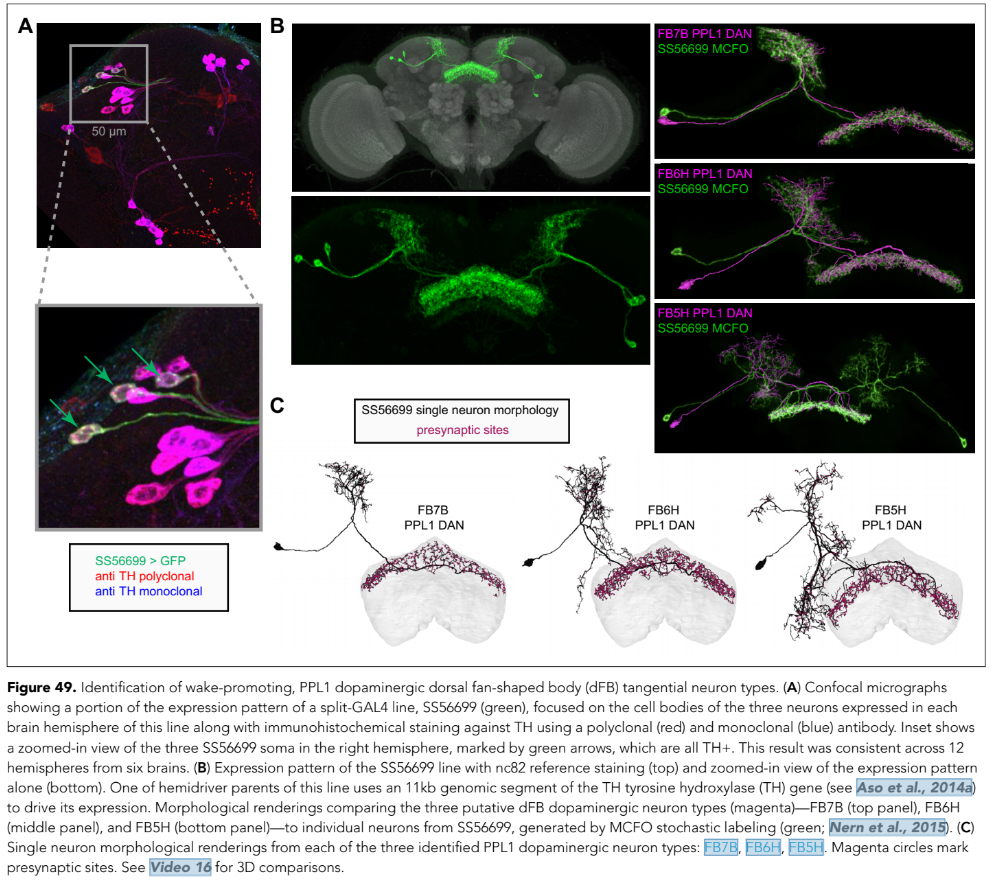
Hulse, B. K., Haberkern, H., Franconville, R., Turner-Evans, D., Takemura, S.-y., Wolff, T., Noorman, M., Dreher, M., Dan, C., Parekh, R., Hermundstad, A. M., Rubin, G. M., & Jayaraman, V. (2021). A connectome of the Drosophila central complex reveals network motifs suitable for flexible navigation and context-dependent action selection. eLife, 10, e66039. https://doi.org/10.7554/eLife.66039
Category: Anatomy, genetics | No Comments
Success: rsh Stock has rsh1 Mutation
on Monday, August 7th, 2023 11:11 | by Isabel Stark
Via gDNA analysis and PCR was the specific area of the rsh gene extracted and amplified where the nucleotide substitution: C to T (Folkers et al., 2006) should be for the rsh1 mutation. The amplicon was Sanger sequenced which proved the nucleotide substitution.
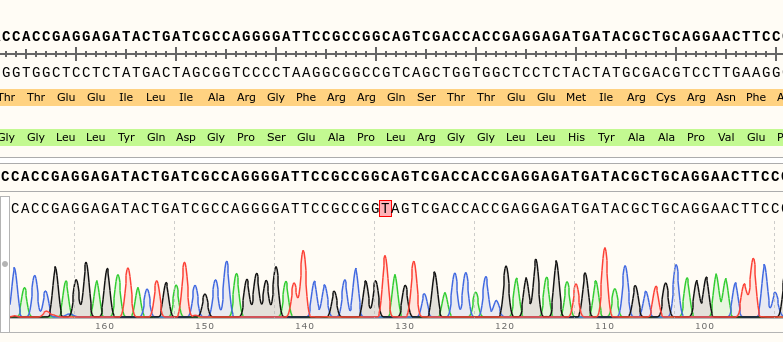
Category: genetics, Memory, Operant learning, operant self-learning, Radish, Uncategorized | No Comments
Creating gRNAs via PCR
on Friday, July 7th, 2023 6:38 | by Isabel Stark
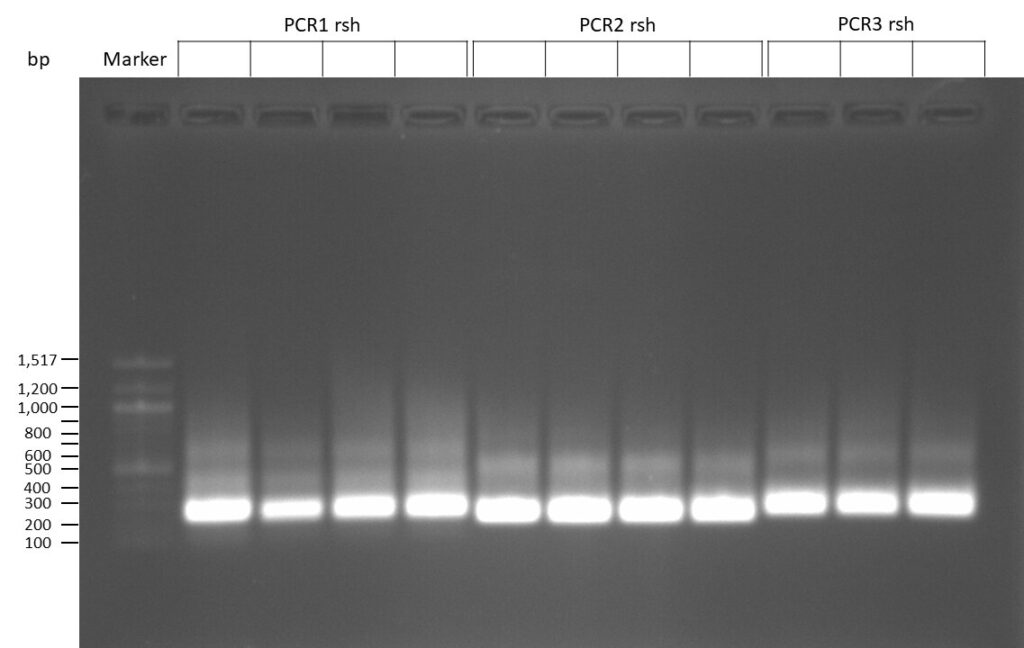
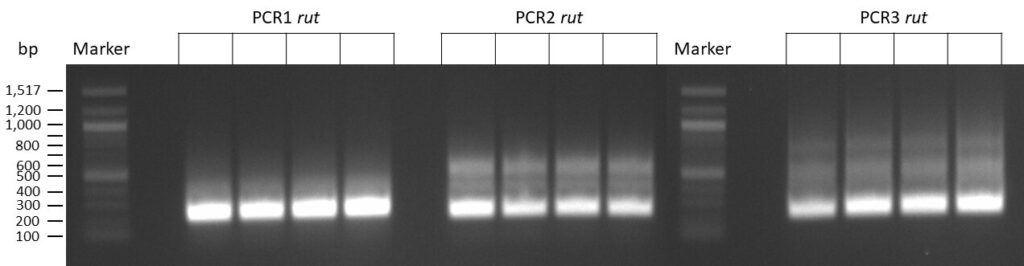
The template pCFD6 was used with a concentration of 640 pg/µl.
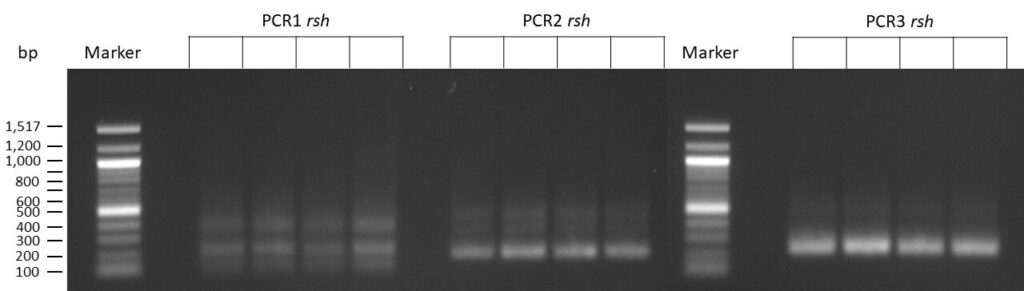
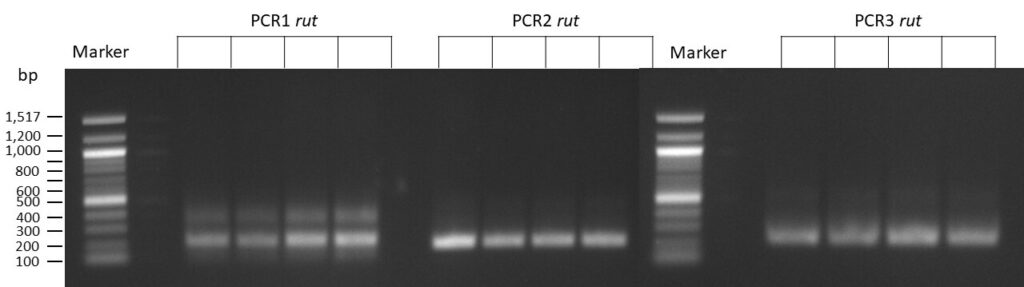
The template pCFD6 was used with a concentration of 64 pg/µl (1:10 dilution).
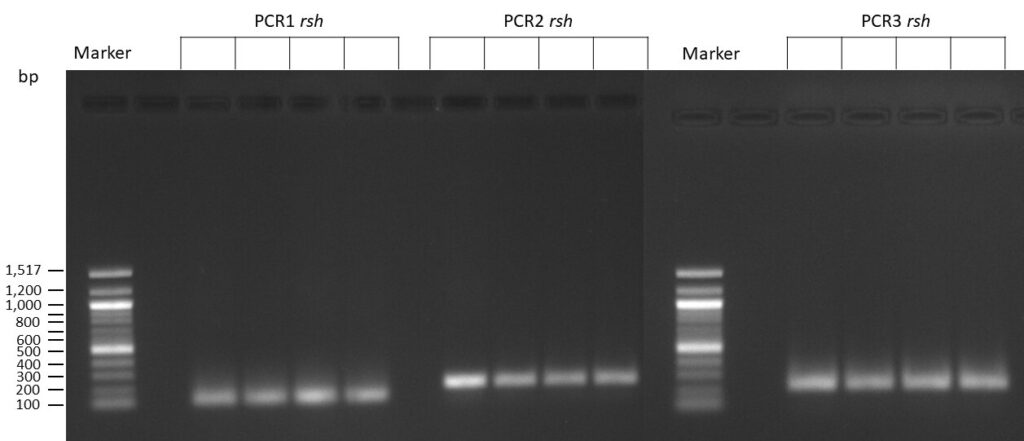
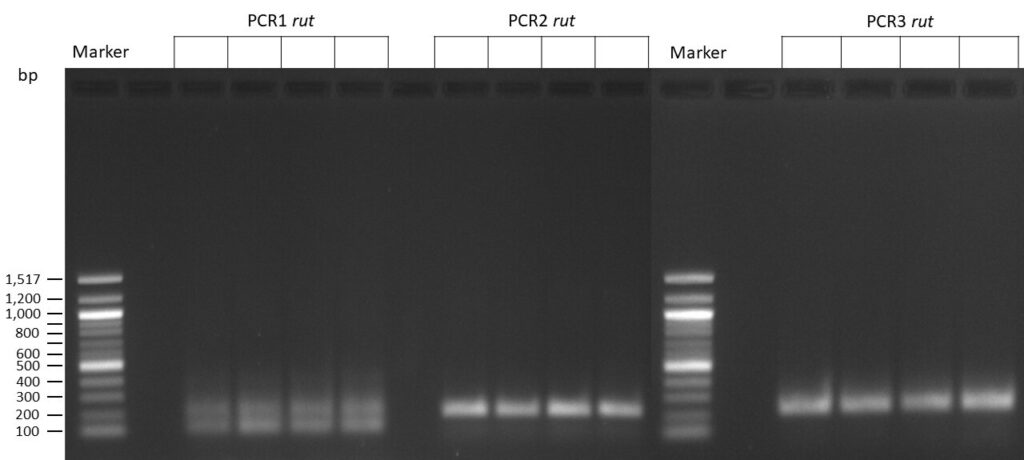
The template pCFD6 was used with a concentration of 64 pg/µl (1:10 dilution).
50µl of 5xQ5 High GC Enhancer was added to the PCR mix.
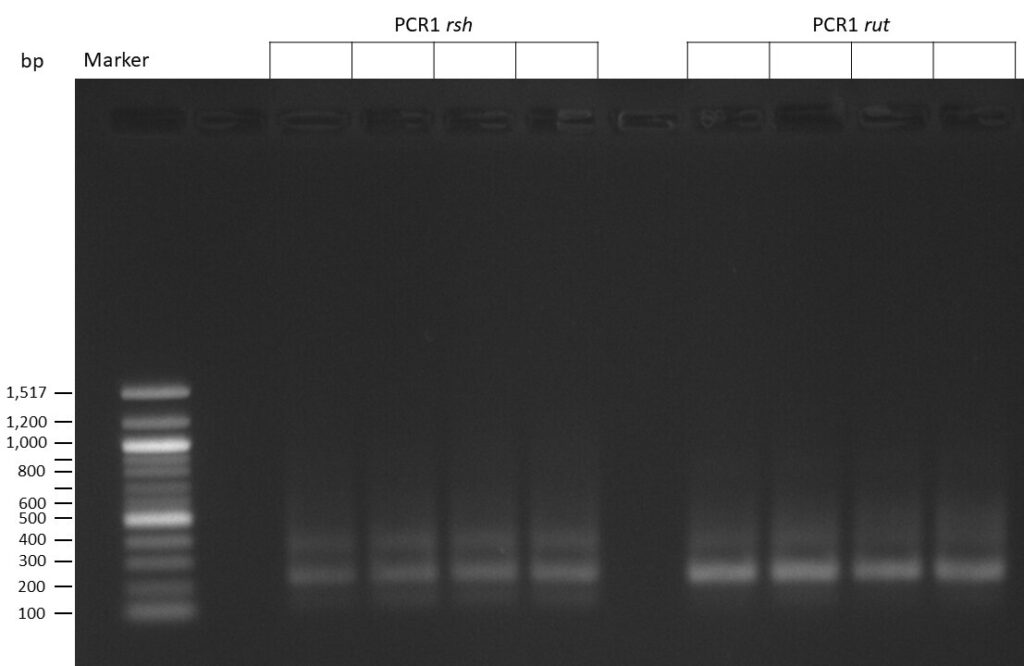
The template pCFD6 was used with a concentration of 128 pg/µl (1:5 dilution).
50µl of 5xQ5 High GC Enhancer was added to the PCR mix.
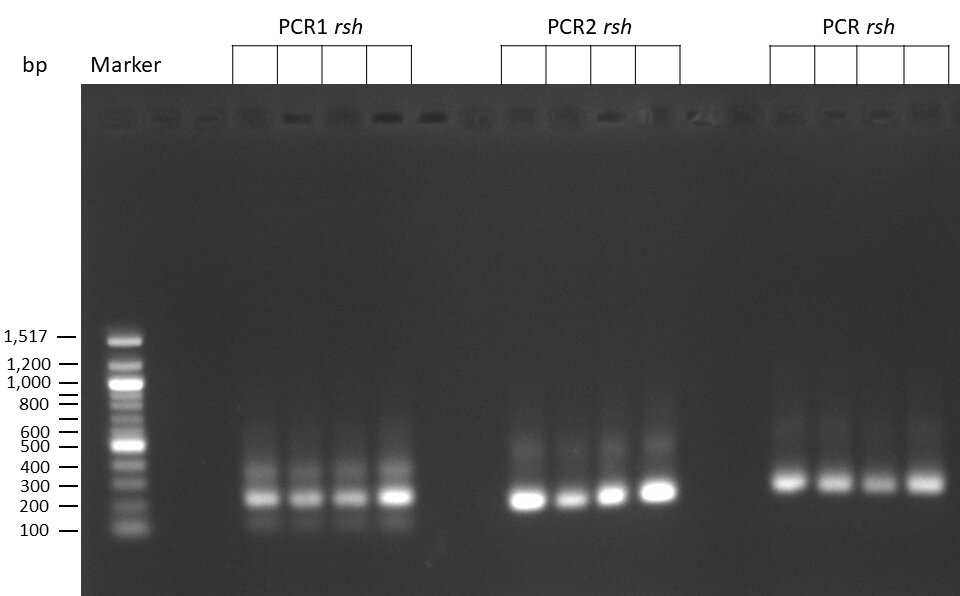
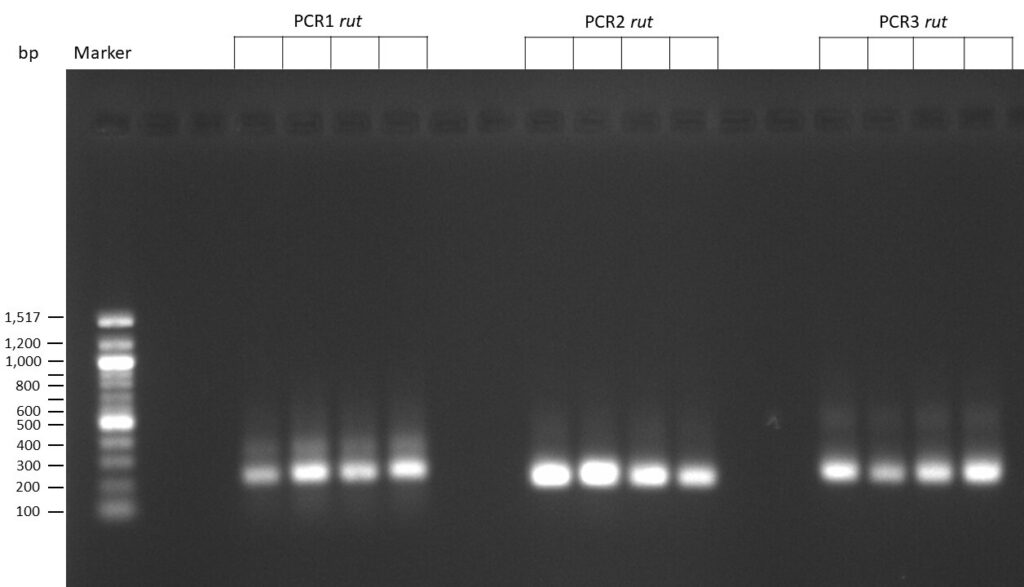
The template pCFD6 was used with a concentration of 640 pg/µl.
The Phusion DNA Polymerase was used instead of the Q5 High-Fidelity DNA Polymerase.
Category: genetics, Memory, Operant learning, operant self-learning, Radish, Uncategorized | No Comments
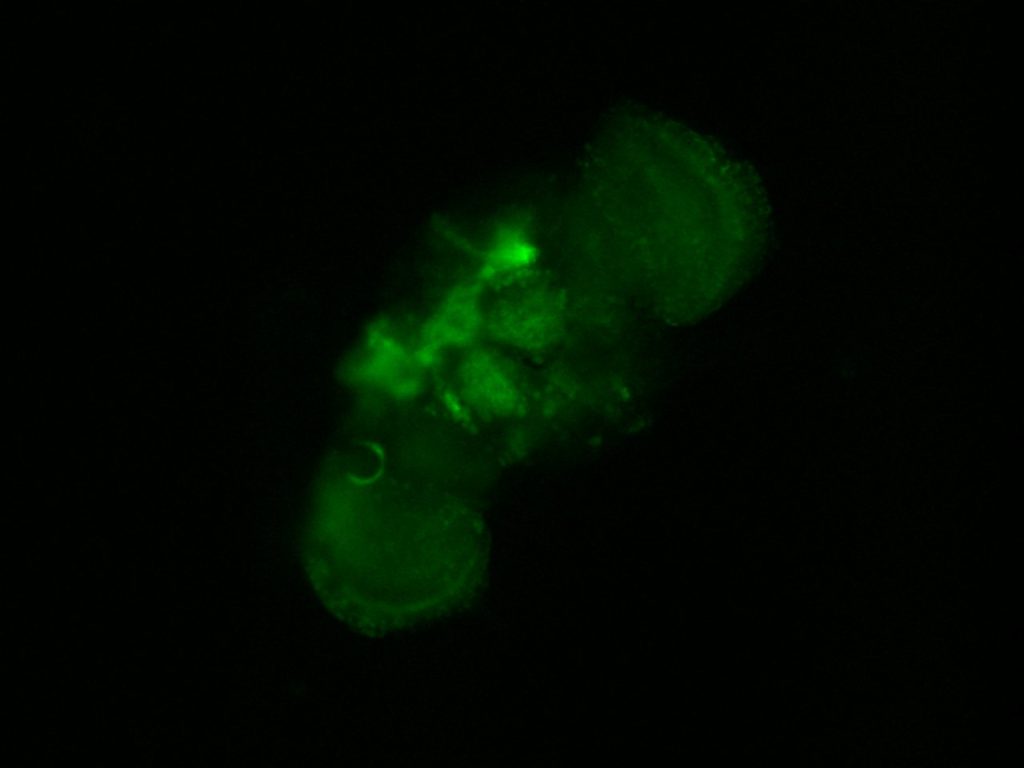
Gene switch finally worked
on Monday, September 28th, 2020 12:21 | by Andreas Ehweiner
I retried the gene-switch method with a new solution of RU486. Since it did not worked before I just tried one line. The flies were left for two days on instant food mixed with a sugar solution of RU486. This time I saw normal GFP expression. So I will test all 4 line in parallel after the break.
Category: Anatomy, crosses | No Comments
