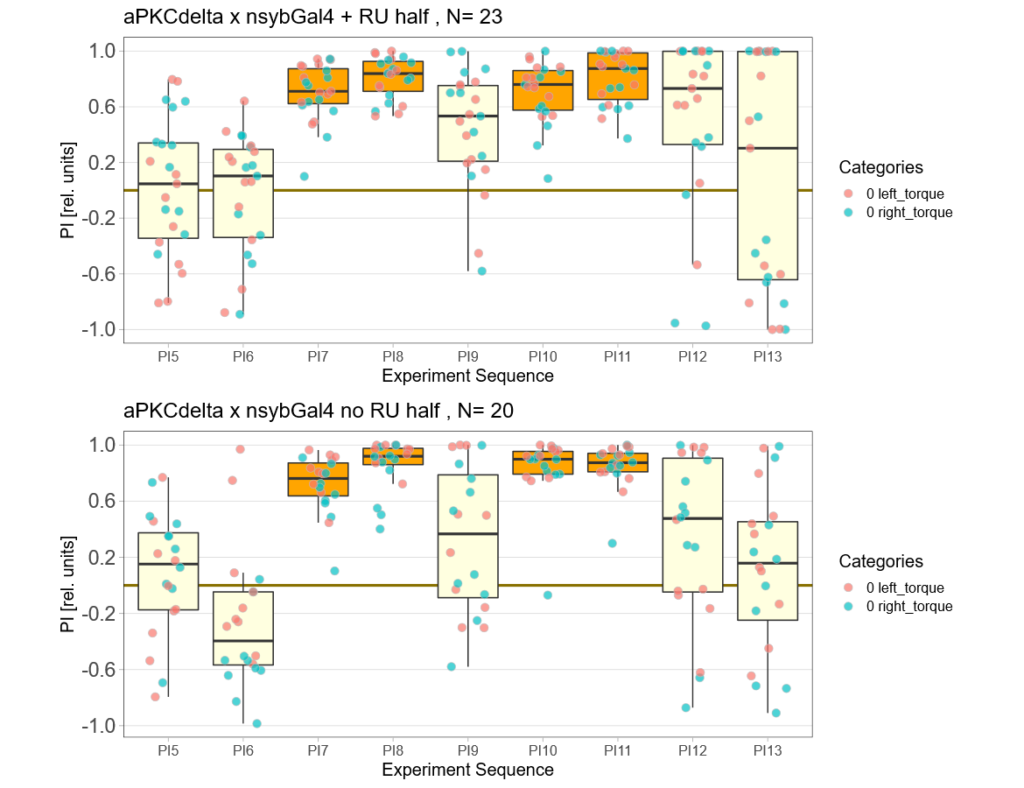
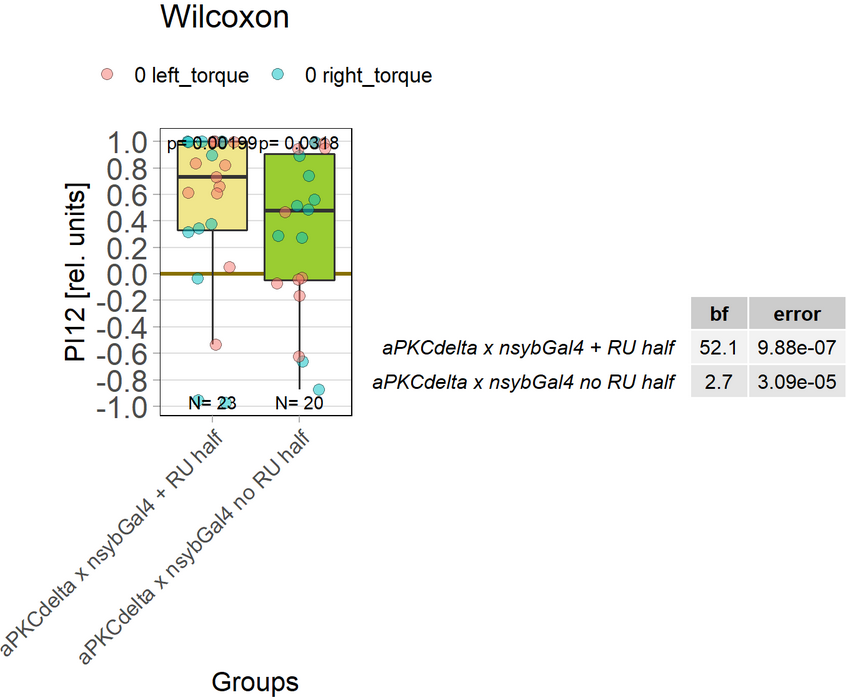
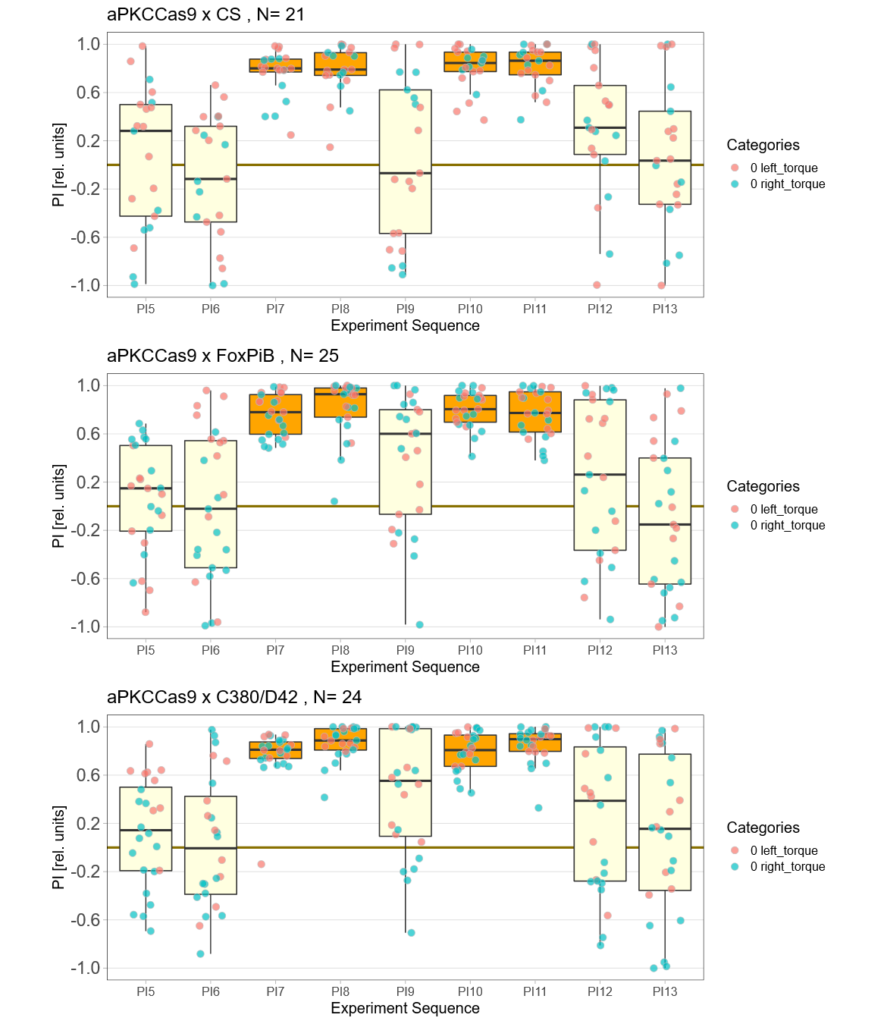
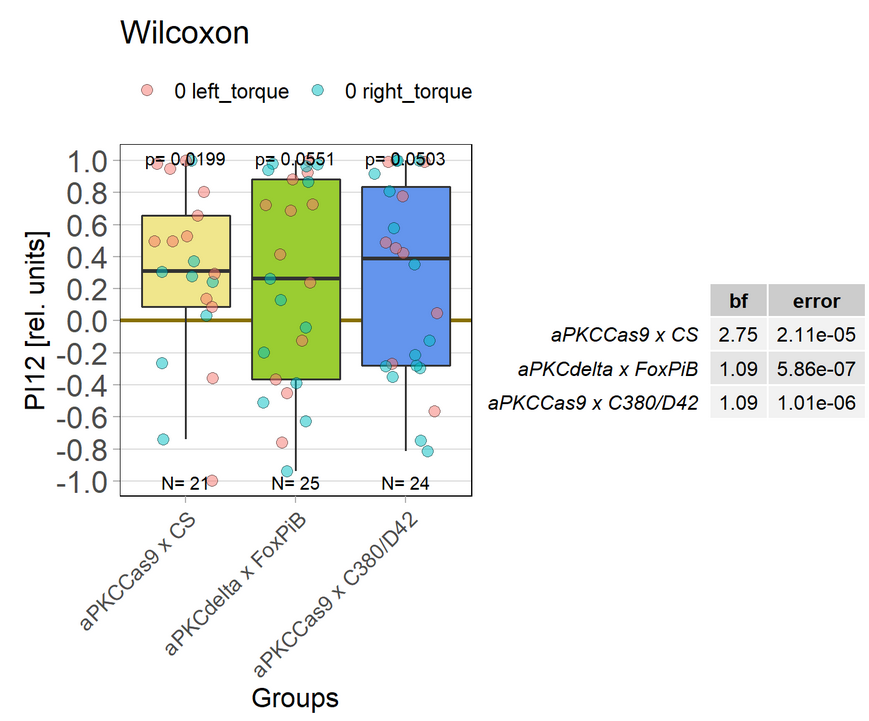
aPKCdelta adult expression half the experiment time
on Monday, December 13th, 2021 9:24 | by Andreas Ehweiner
Expression of aPKCdelta in all neurons in the adult fly (nysbGS-Gal4) for half the experiment time (1 min periods).
Category: flight, Memory, Operant learning, operant self-learning, PKC, Uncategorized | No Comments
The reaction of drosophila to light
on Friday, December 3rd, 2021 9:35 | by Parva Nasimi
NorpA-UAS-Chrimson/66009,
NorpA-UAS-Chrimson/66555,
NorpA-UAS-Chrimson/38764,
NorpA-UAS-Chrimson/0273, and NorpA-UAS-Chrimson/66010 flies to white light (ATR-treated foods for 2days).
Category: Uncategorized | 1 Comment
aPKCdelta half the experiment time
on Monday, November 8th, 2021 10:49 | by Andreas Ehweiner
Expression of aPKCdelta in FoxPiB positiv or motor neurons, the time of the experiment is half of the normal time.
Category: flight, Foxp, Memory, Operant learning, operant self-learning, PKC, Uncategorized | No Comments
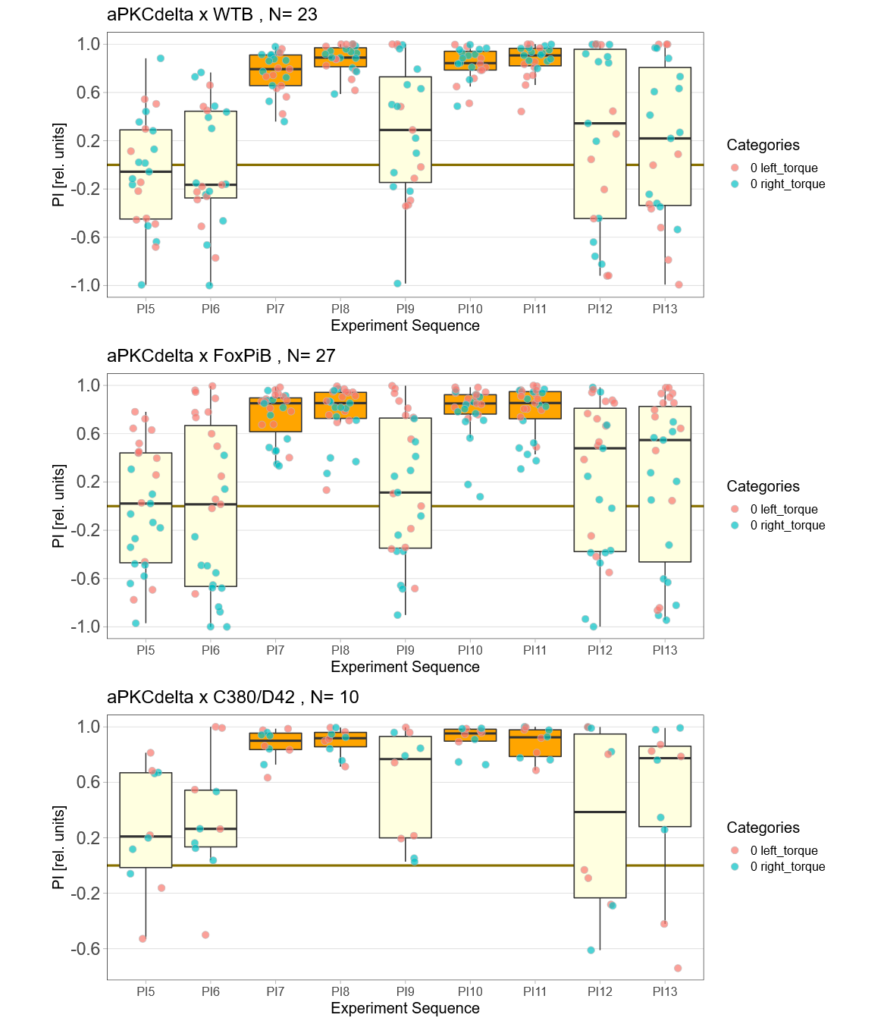
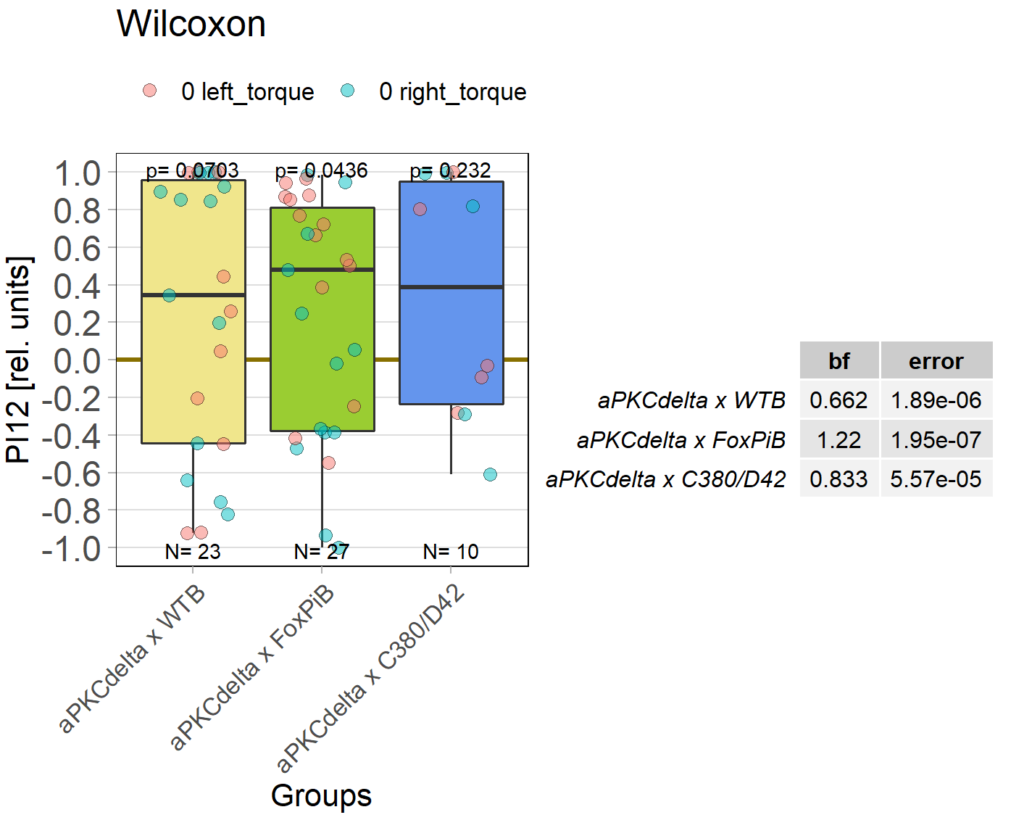
aPKCdelta expression
on Monday, October 25th, 2021 12:01 | by Andreas Ehweiner
Category: flight, Foxp, Operant learning, operant self-learning, PKC, Uncategorized | No Comments
FasII Immunohistochemistry
on Sunday, October 10th, 2021 10:49 | by Parva Nasimi
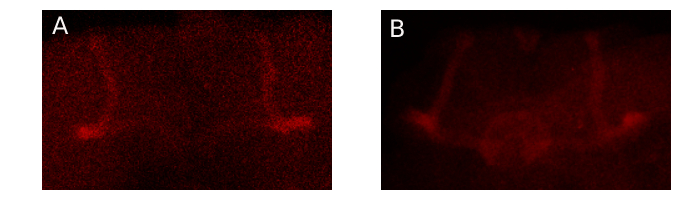
FasII staining:
Mushrom bodies of A: Control, B: Foxp ko.
Category: Uncategorized | No Comments
T-Maze experiment with Red Light
on Monday, September 20th, 2021 1:05 | by Parva Nasimi
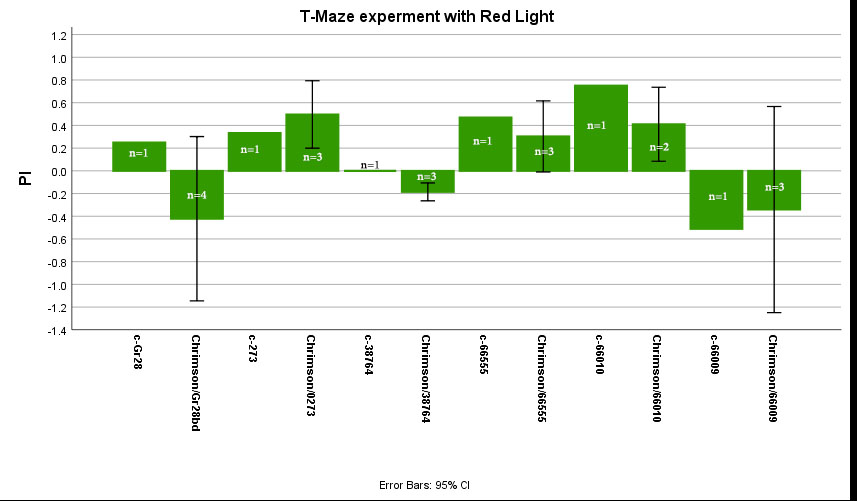
T-Maze experiment with Red light:
Graph shows “Mean Performance Index” for NorpA-UAS-Chrimson/Gr28bd, NorpA-UAS-Chrimson/0273, NorpA-UAS-Chrimson/38764, NorpA-UAS-Chrimson/66555, NorpA-UAS-Chrimson/66010, NorpA-UAS-Chrimson/66009 (ATR-treated foods for 2days) and Controls (c-= without ATR).
Category: Uncategorized | No Comments
T-Maze experiment with yellow and red lights
on Tuesday, September 7th, 2021 8:40 | by Parva Nasimi
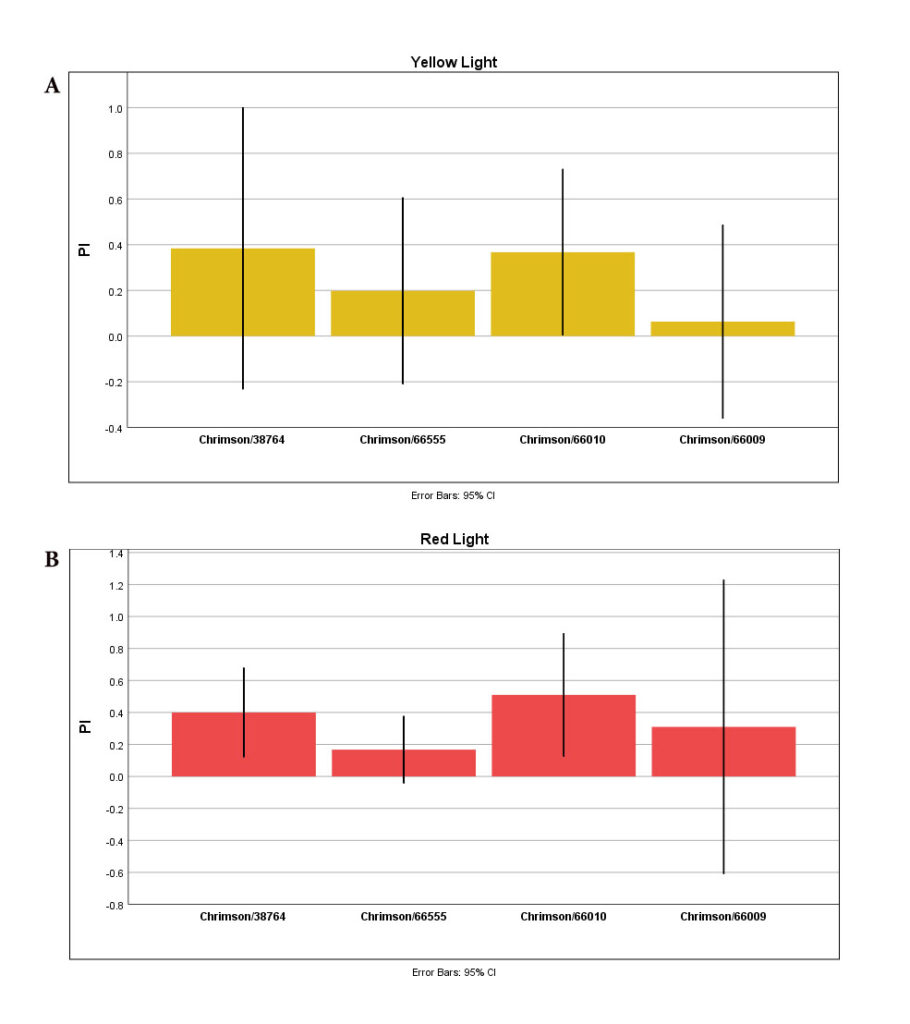
T-Maze experiment with yellow (A) and red (B) lights:
Graphs show “Mean Performance Index” for NorpA-UAS-Chrimson/38764, NorpA-UAS-Chrimson/66555, NorpA-UAS-Chrimson/66010, and NorpA-UAS-Chrimson/66009.
Flies (n=3) were given ATR-treated foods for 2days.
The same flies were used for both experiments, first with red light and the next day with yellow light.
Category: Uncategorized | No Comments
T-Maze experiment with red light
on Tuesday, September 7th, 2021 12:02 | by Parva Nasimi

T-Maze experiment with red light.
Graph shows “Mean Performance Index” for NorpA-UAS-Chrimson/Gr28bd, NorpA-UAS-Chrimson/0273-Gal4, NorpA-UAS-Chrimson/38764, NorpA-UAS-Chrimson/66555, NorpA-UAS-Chrimson/66010, and NorpA-UAS-Chrimson/66009.
Flies were given ATR-treated foods for 2days.
Category: Uncategorized | No Comments
Immunostaining of Mushroom Body using anti-FasII antibody
on Wednesday, September 1st, 2021 11:36 | by Parva Nasimi
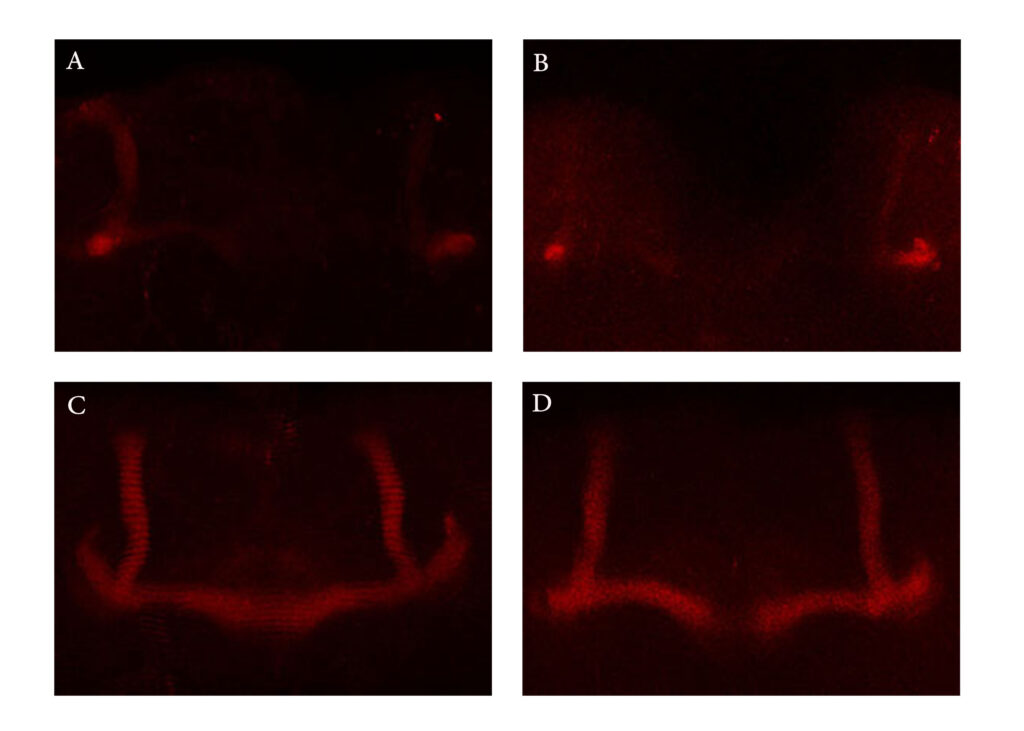
Category: Uncategorized | No Comments
Colocalization of FoxP
on Monday, August 30th, 2021 12:48 | by Andreas Ehweiner
Cross of FoxPLexA (red) with diffrent GFP-Lines (green)
GMR65A06
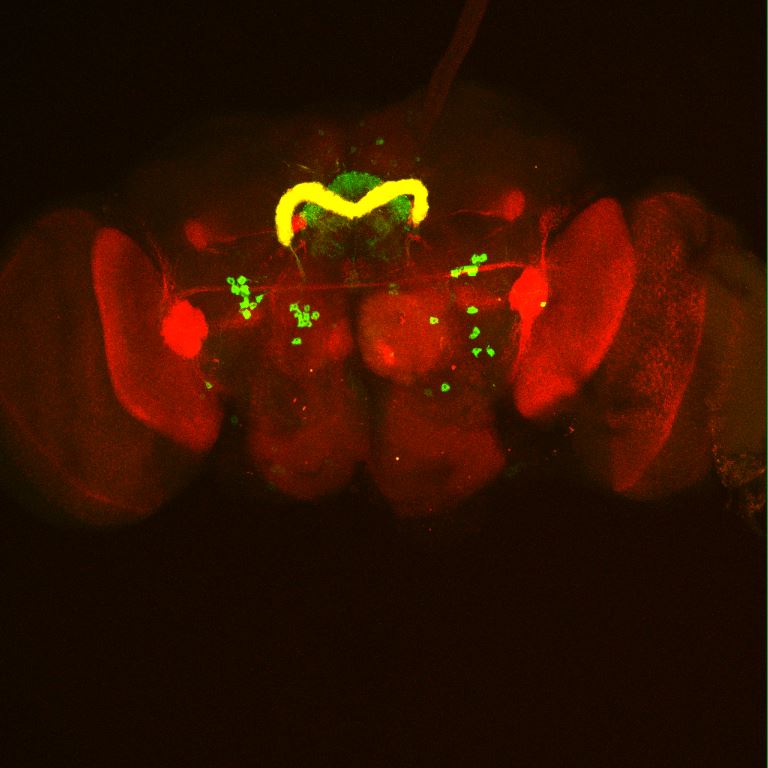
GMR20H05
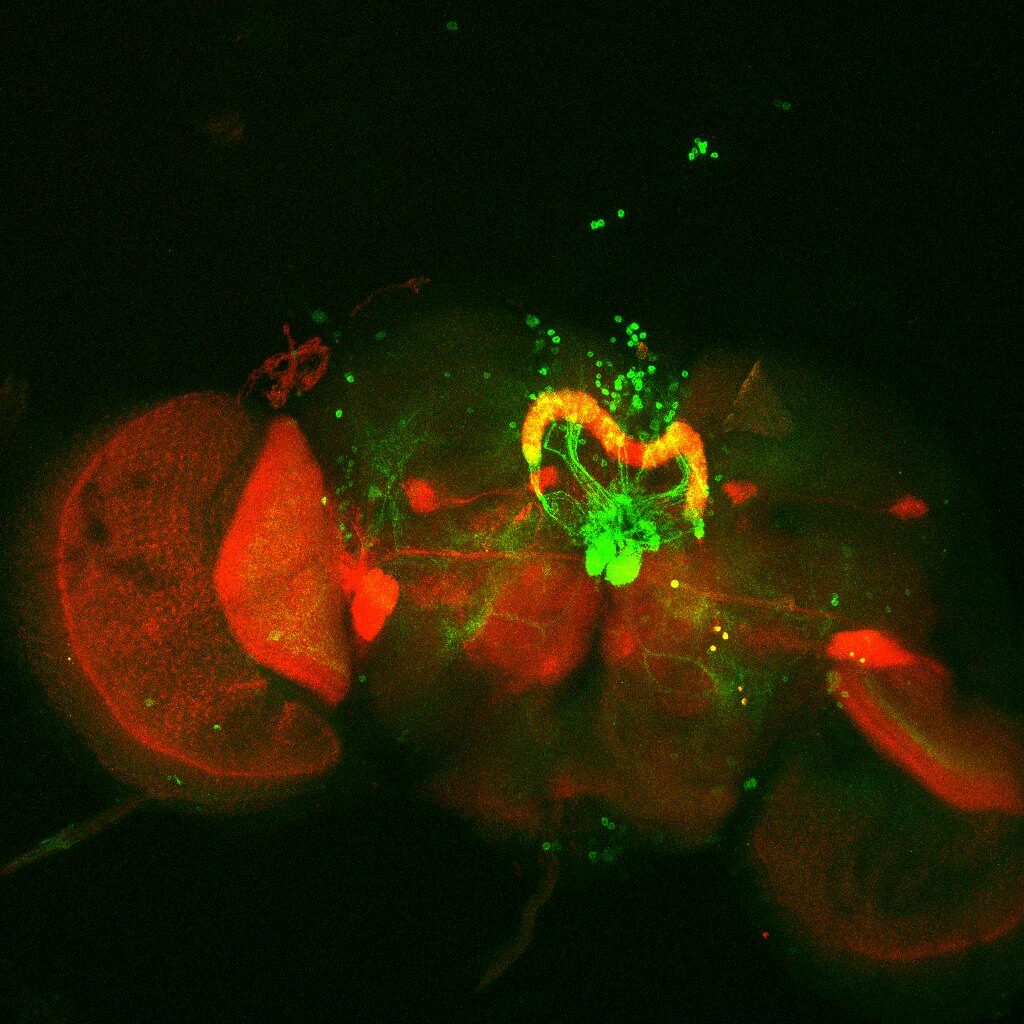
GMR11F02 (dose not overlap)
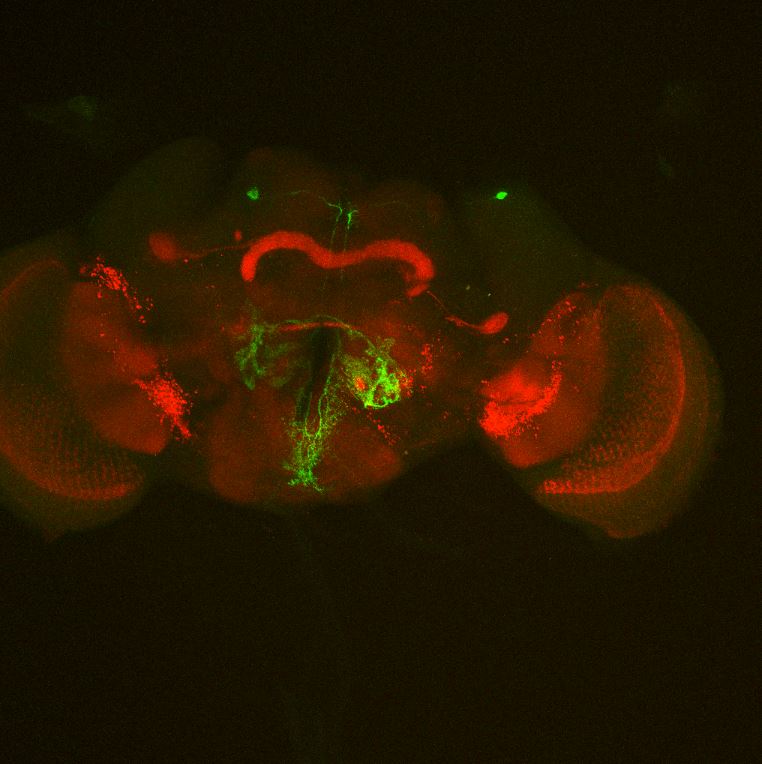
GMR52B10 not checked yet
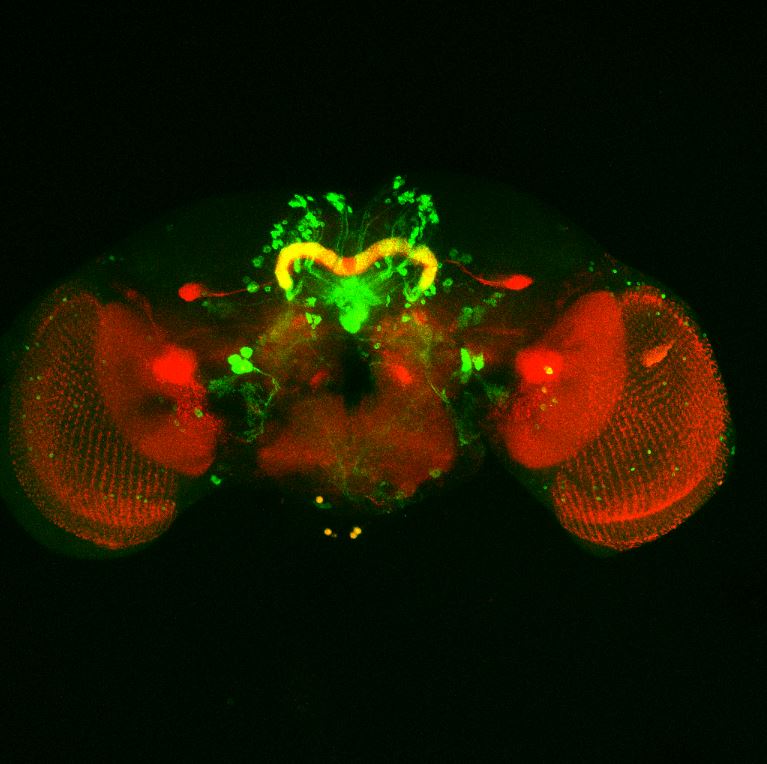
aPKC (must be repeated)
Category: Anatomy, Foxp, Uncategorized | No Comments