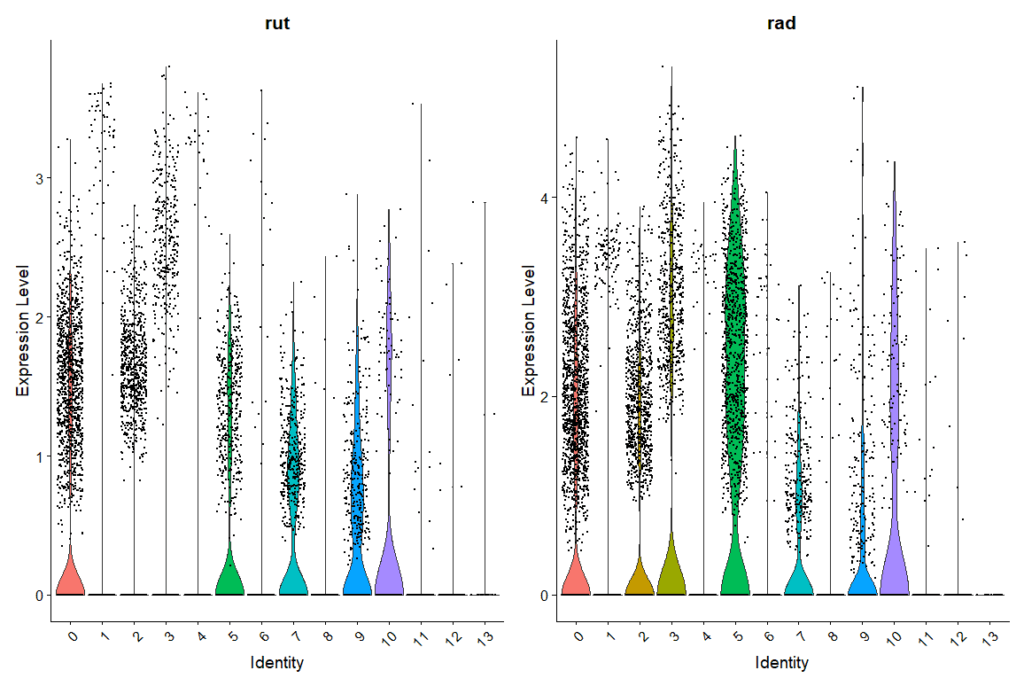
Rut and Rad Expression in Ventral Nerve Cord
on Monday, January 12th, 2026 1:08 | by Ipek Subay
It will be updated with the source of the dataset
Category: Uncategorized | No Comments
Torquemeter Test and Confocal images of MBON02
on Monday, December 22nd, 2025 10:08 | by Ipek Subay
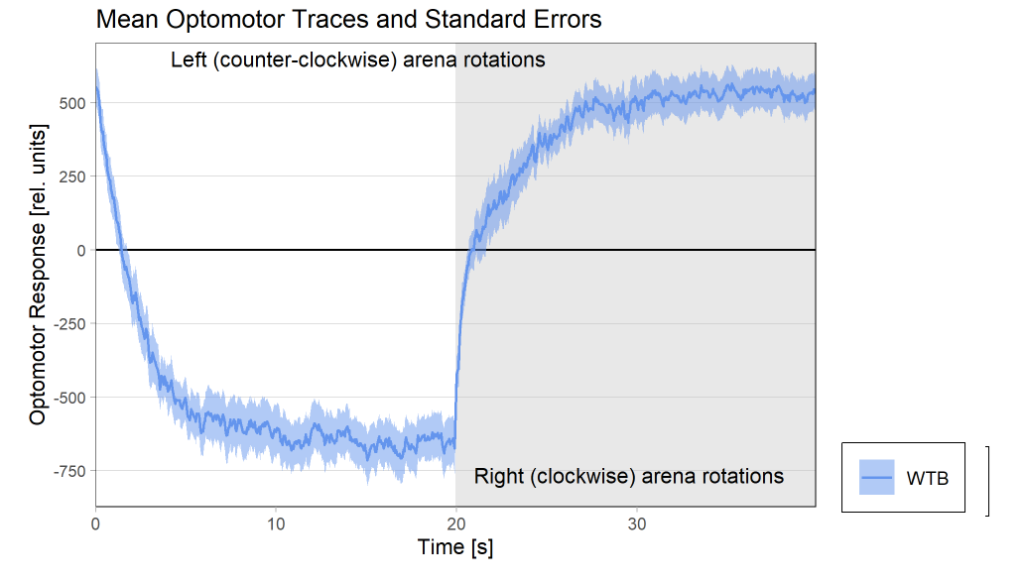
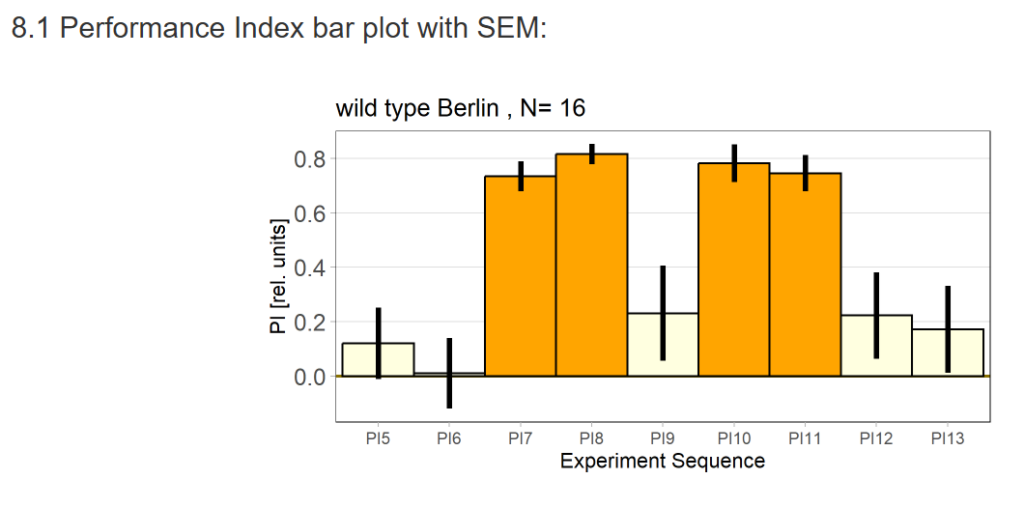
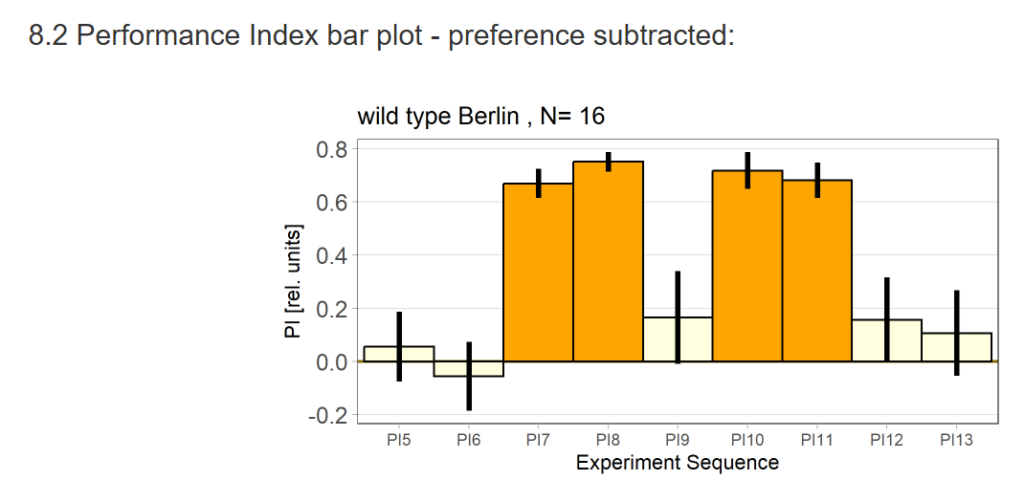
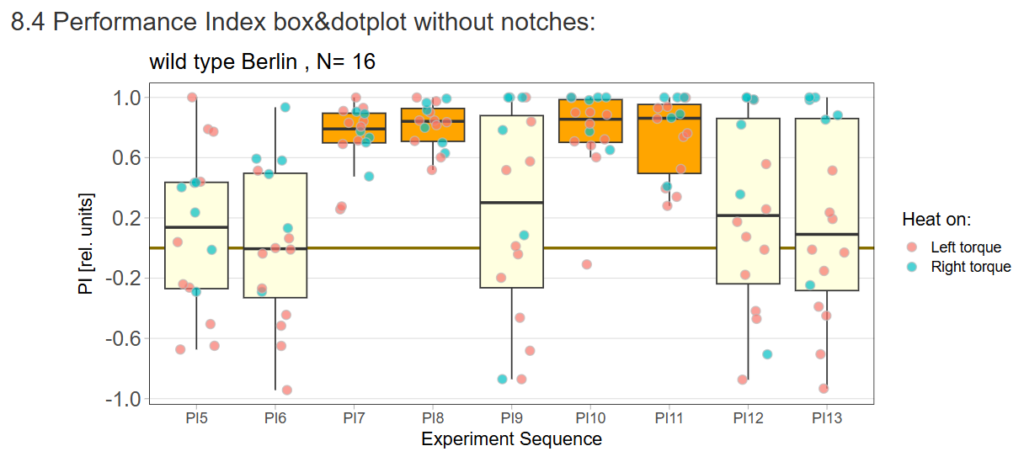
16 flies went through the test out of 48 flies
I retook the images of last week’s brains for MBON02-transtango (trans-tango mkII) imaging. Green channel shows MBON02, red channel shows post-synaptic cells of MBON02:
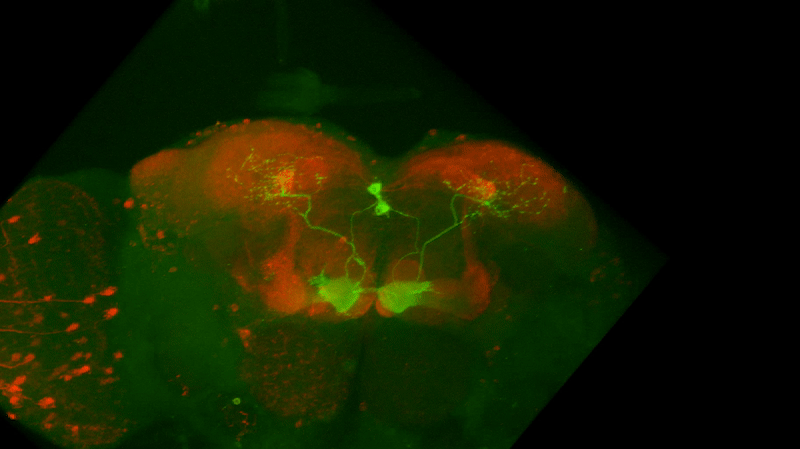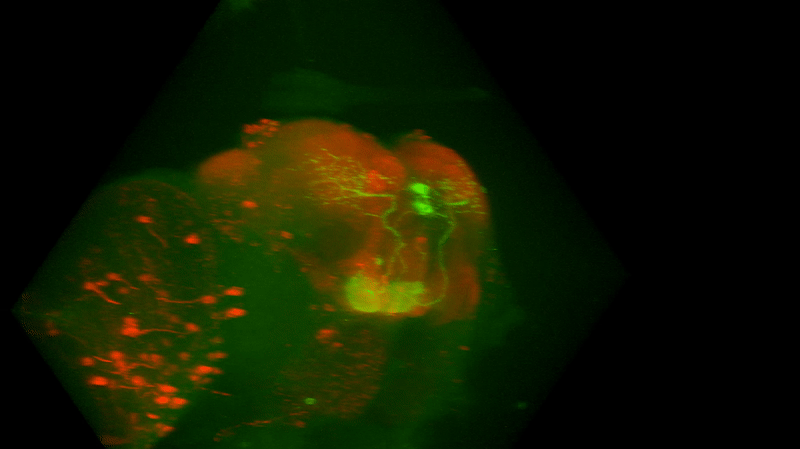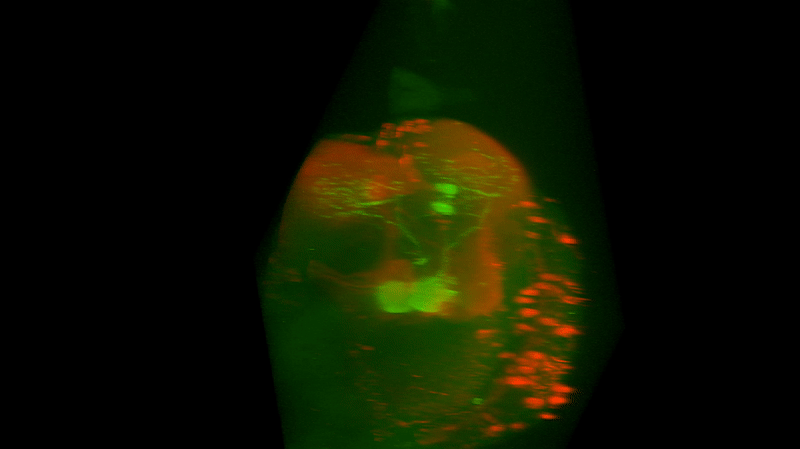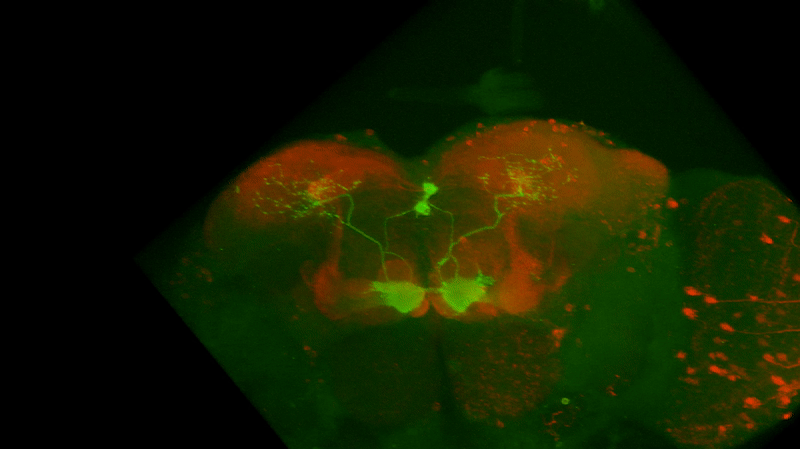
Category: Uncategorized | No Comments
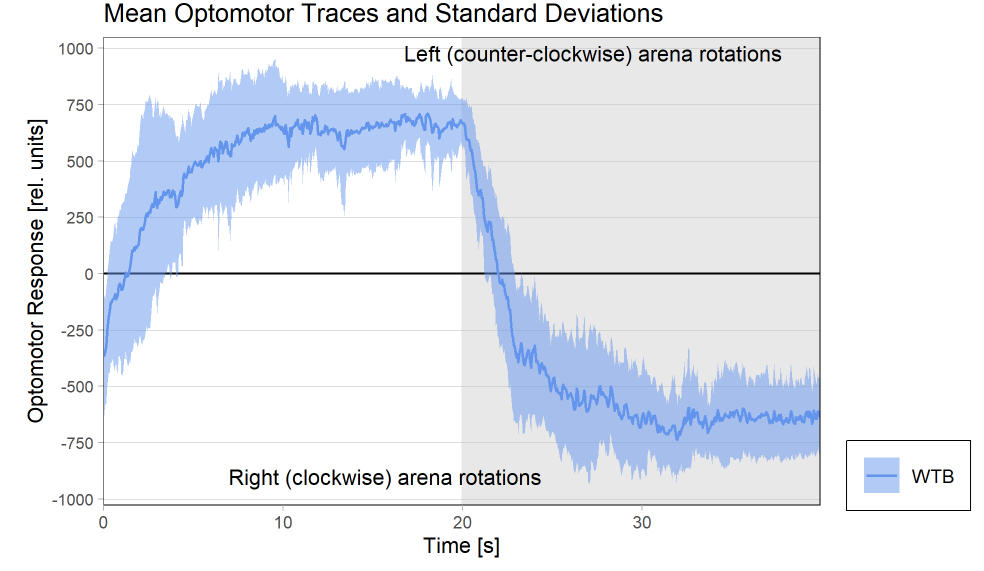
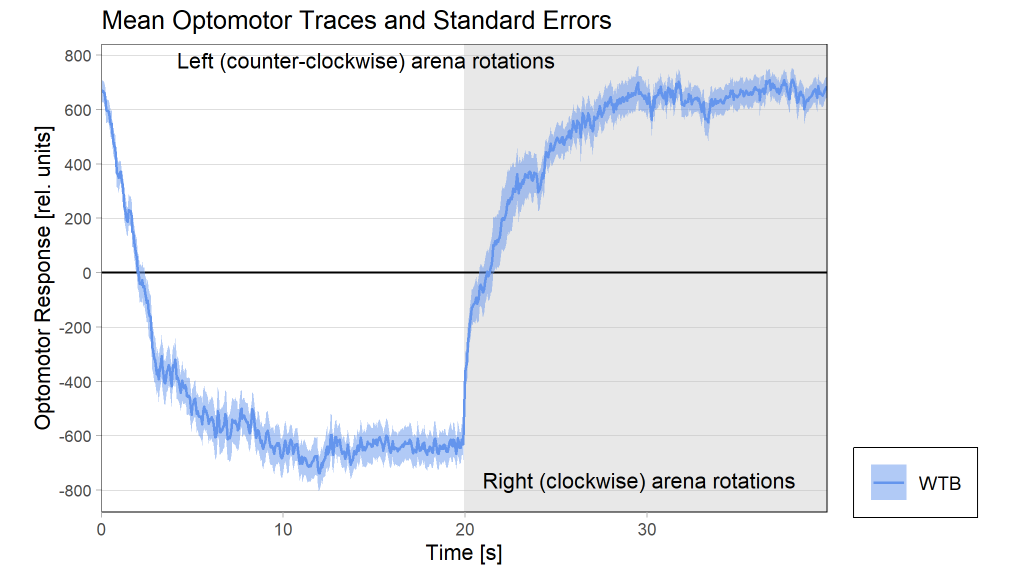
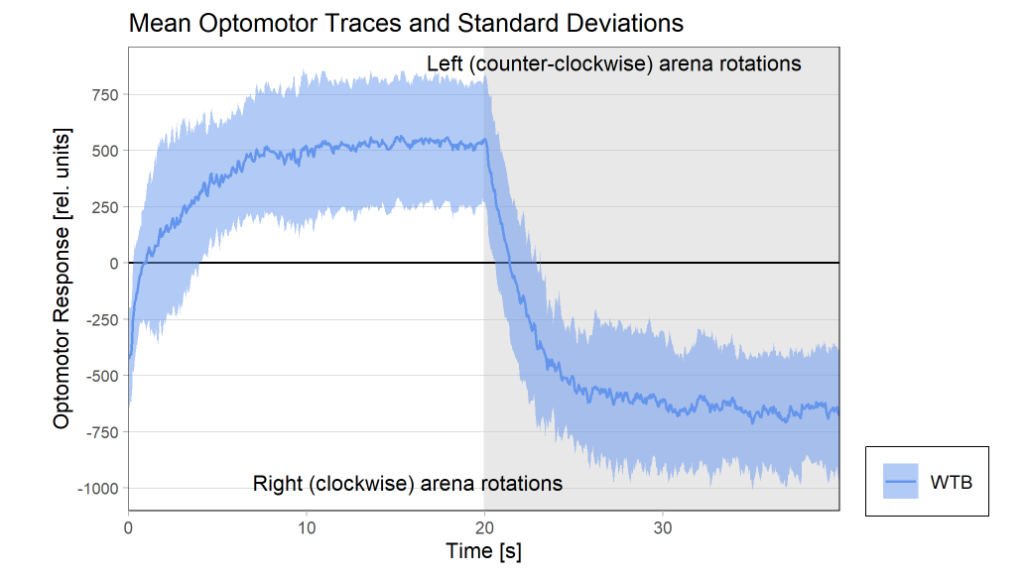
Results torque meter 18th and 19th
on Monday, November 24th, 2025 12:46 | by Fridrik Kjartansson
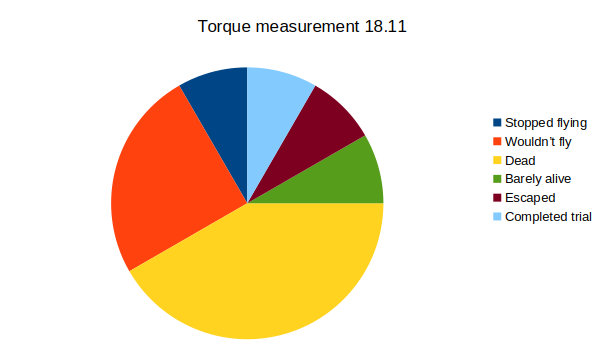
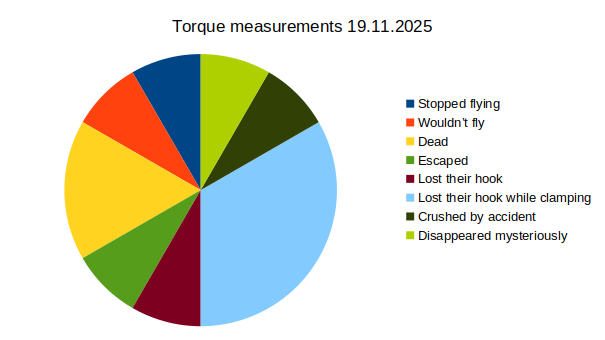
Category: Uncategorized | No Comments
Confocal images 21.11.2025 powerpoint
on Monday, November 24th, 2025 10:51 | by Fridrik Kjartansson
Category: Uncategorized | No Comments
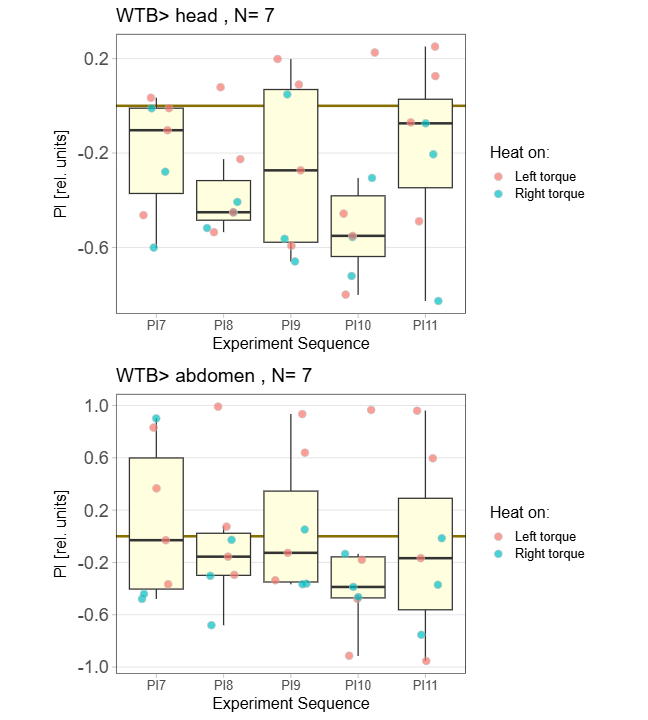
WTB Learning Test
on Monday, November 24th, 2025 10:47 | by Ipek Subay

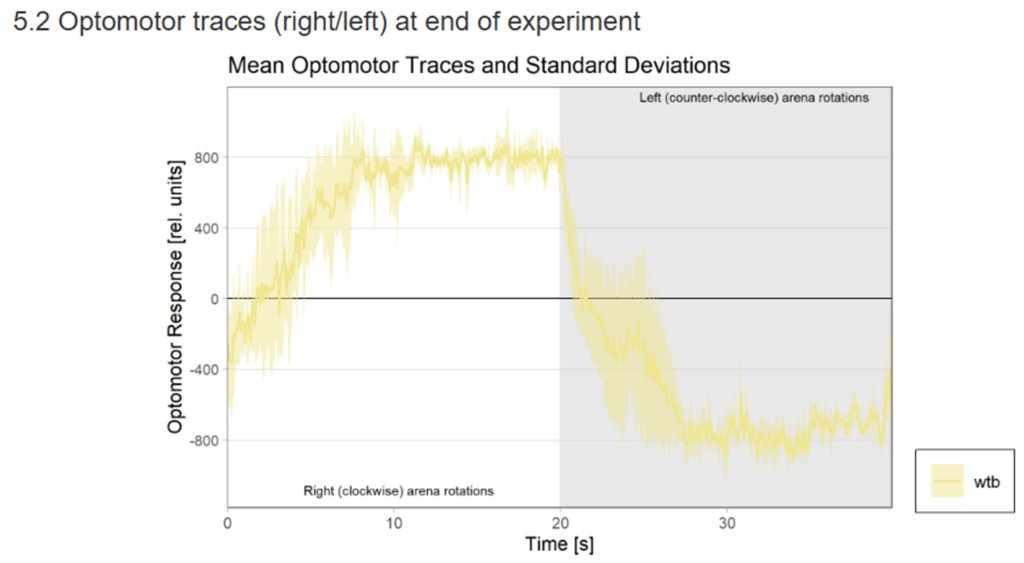
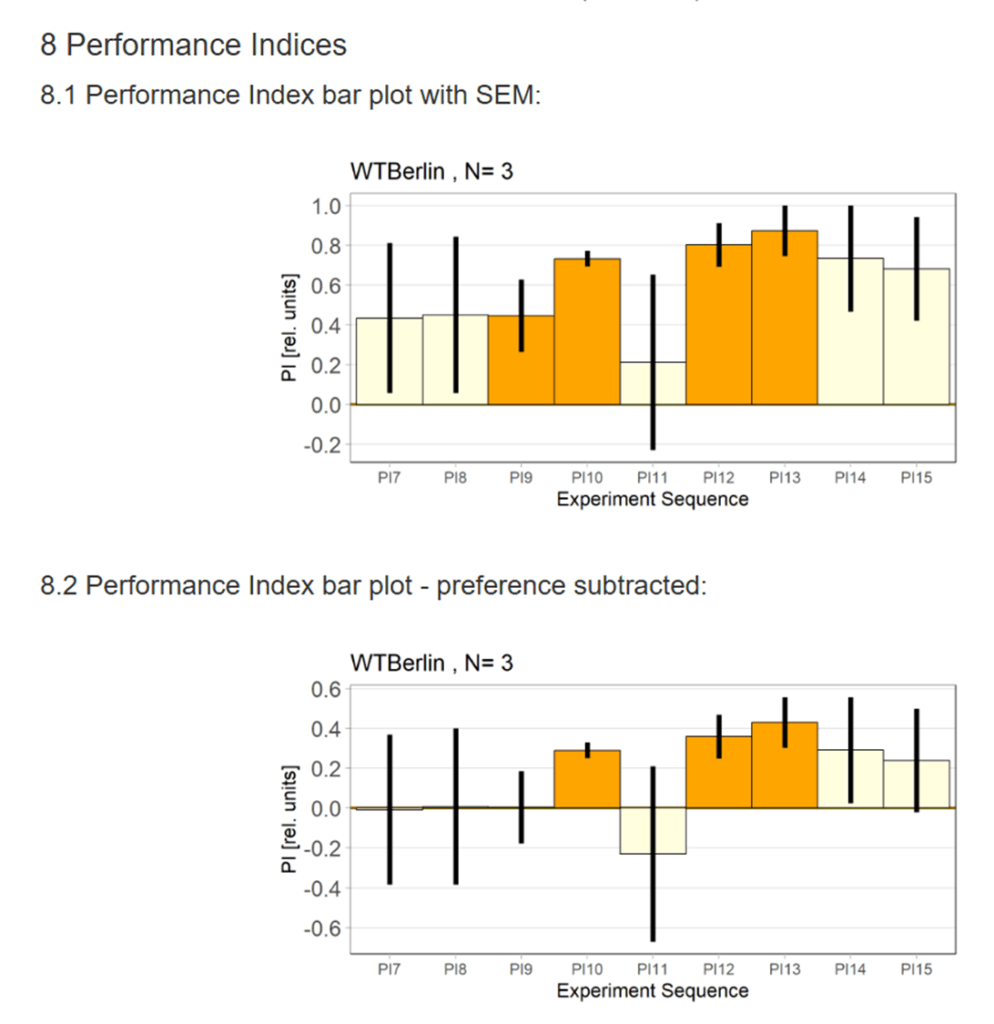
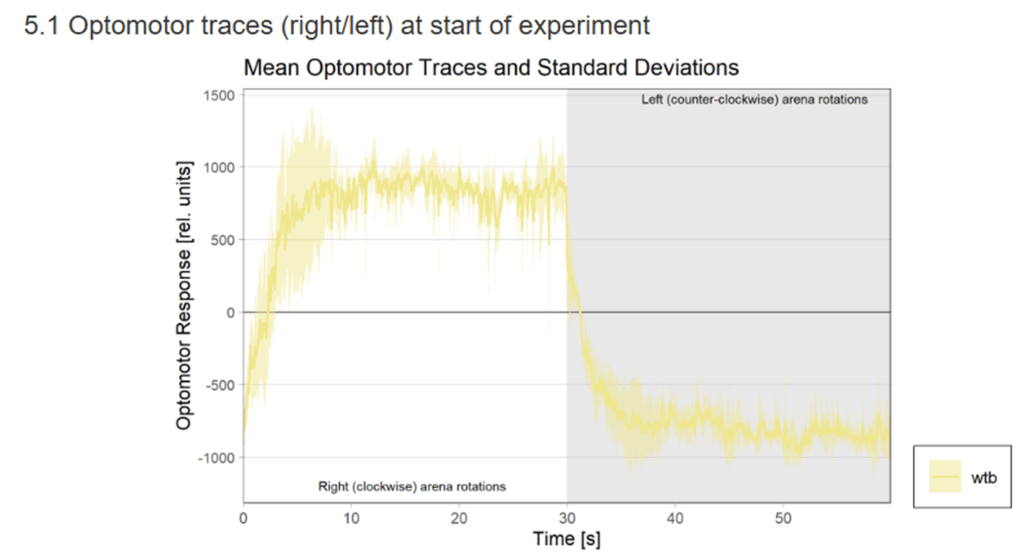
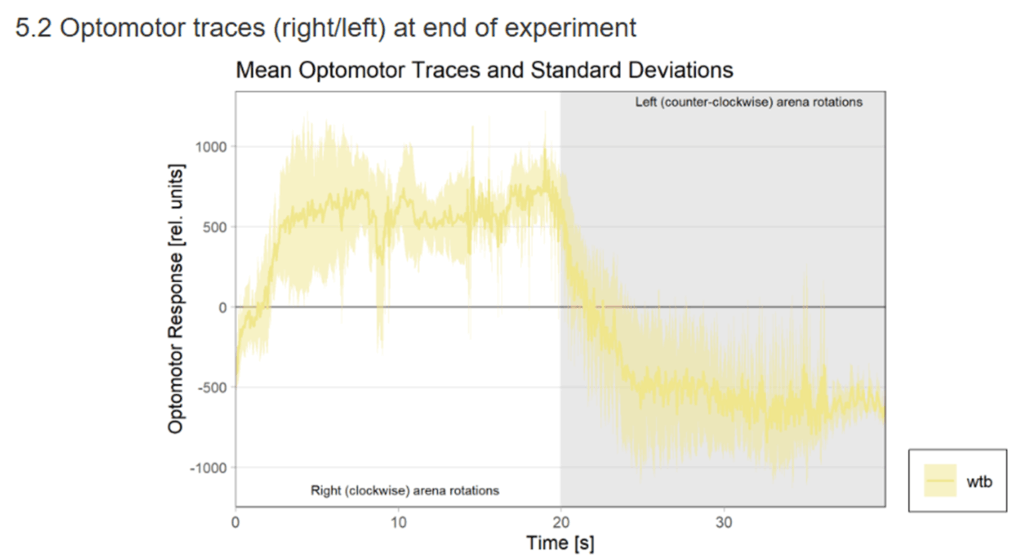
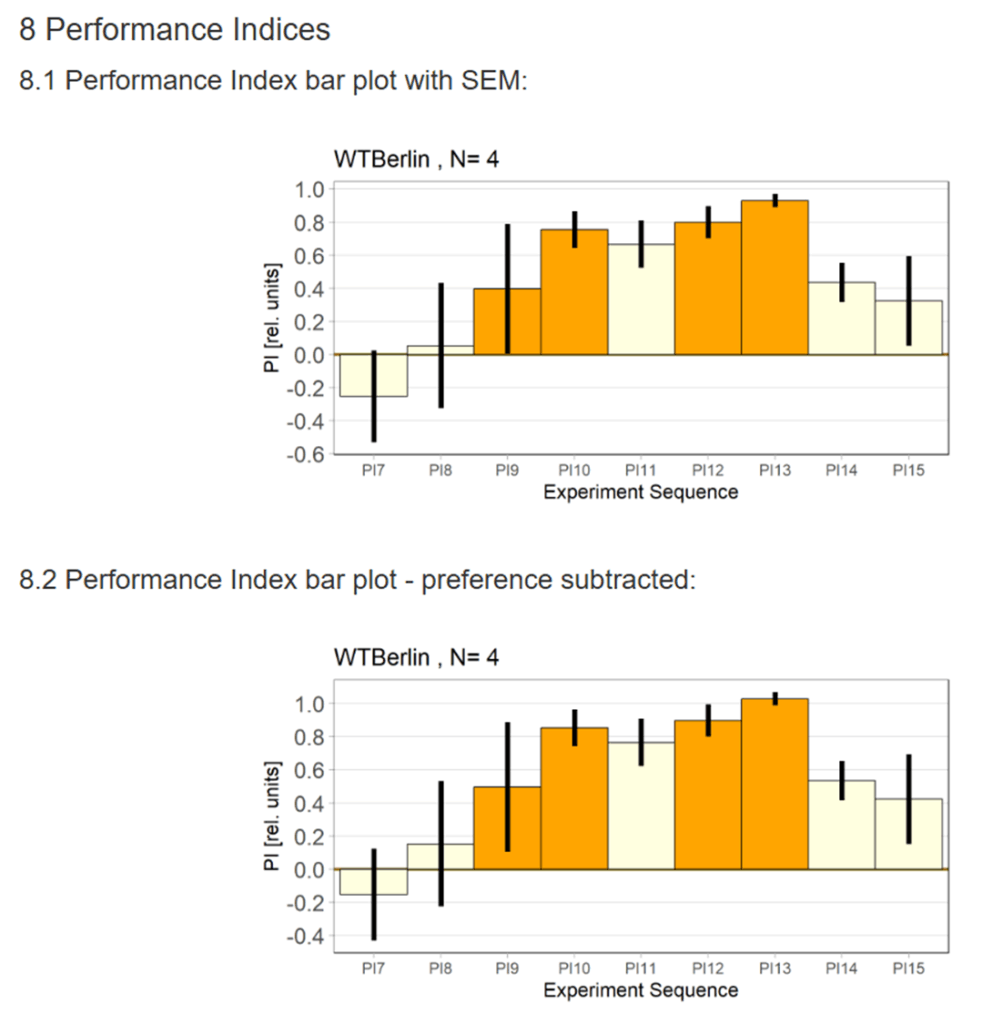
Category: Uncategorized | No Comments
Confocal images
on Monday, November 24th, 2025 10:20 | by Fridrik Kjartansson
Category: Uncategorized | No Comments
Work flow molecular work
on Monday, October 27th, 2025 1:03 | by Julia Schulz
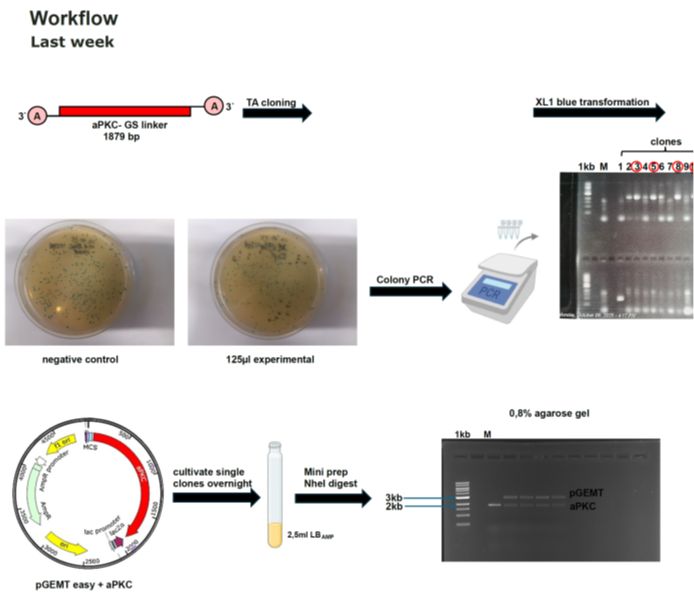
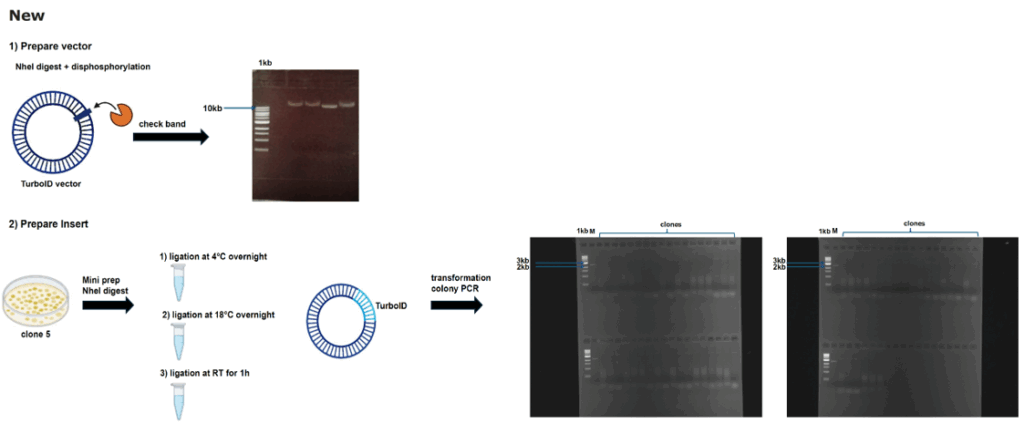
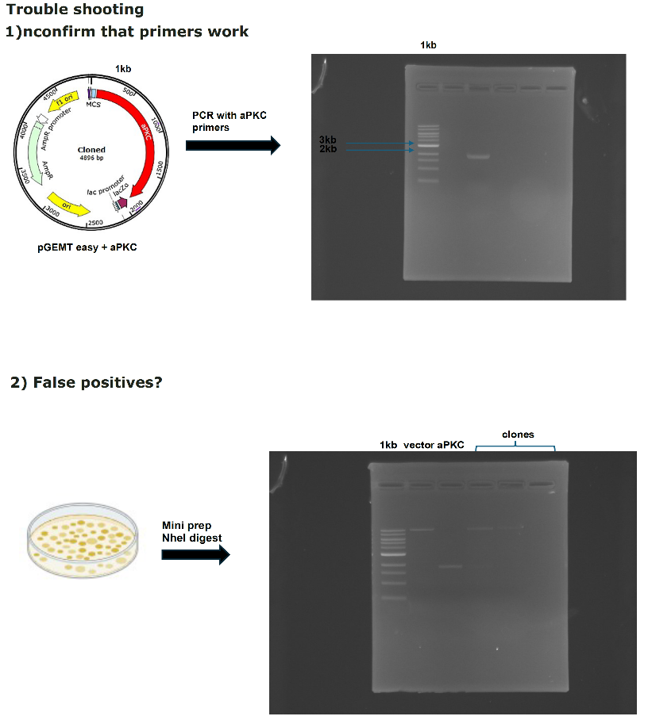
Category: Uncategorized | No Comments
WTB fly, first test
on Monday, October 20th, 2025 12:21 | by Fridrik Kjartansson
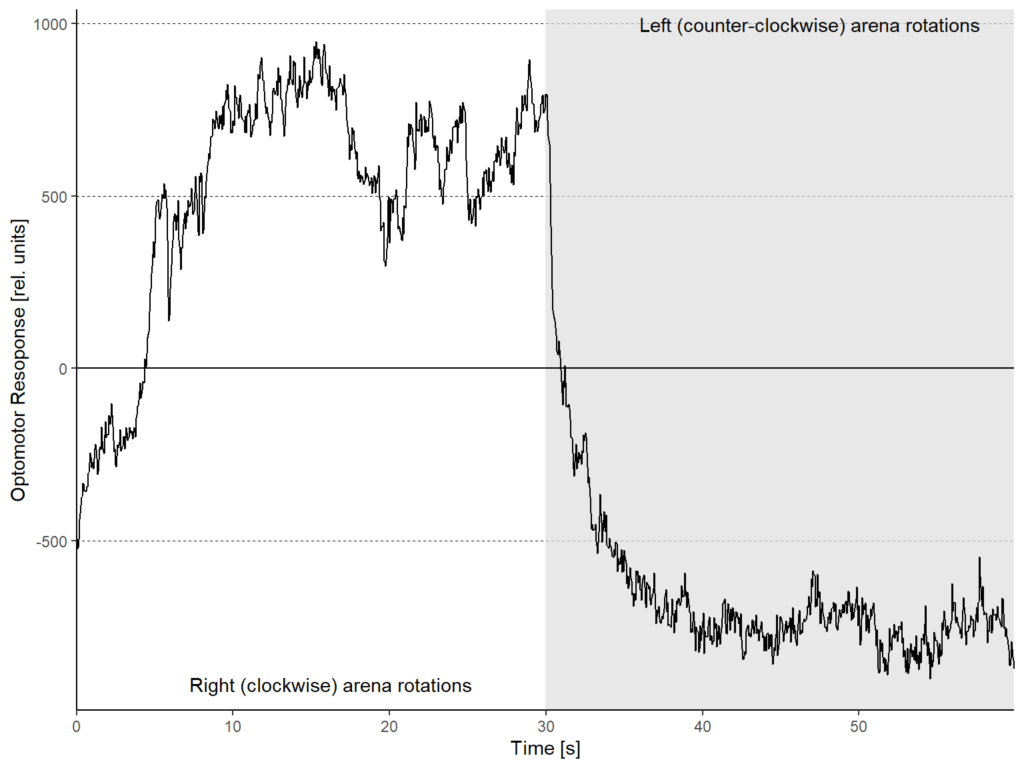
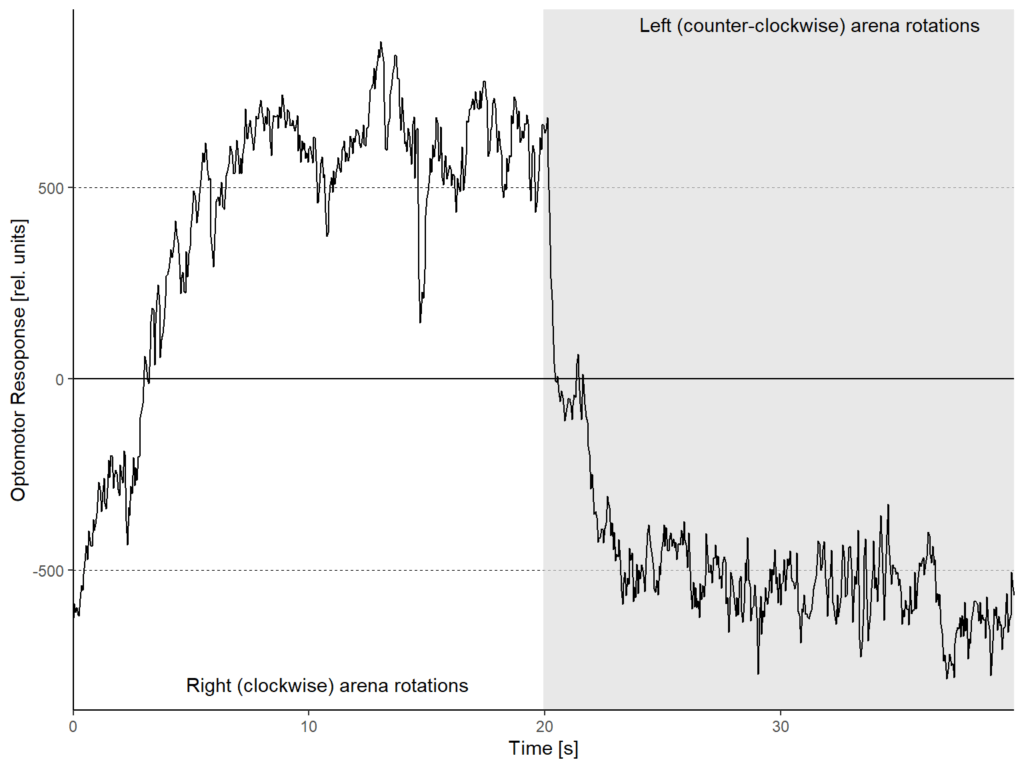
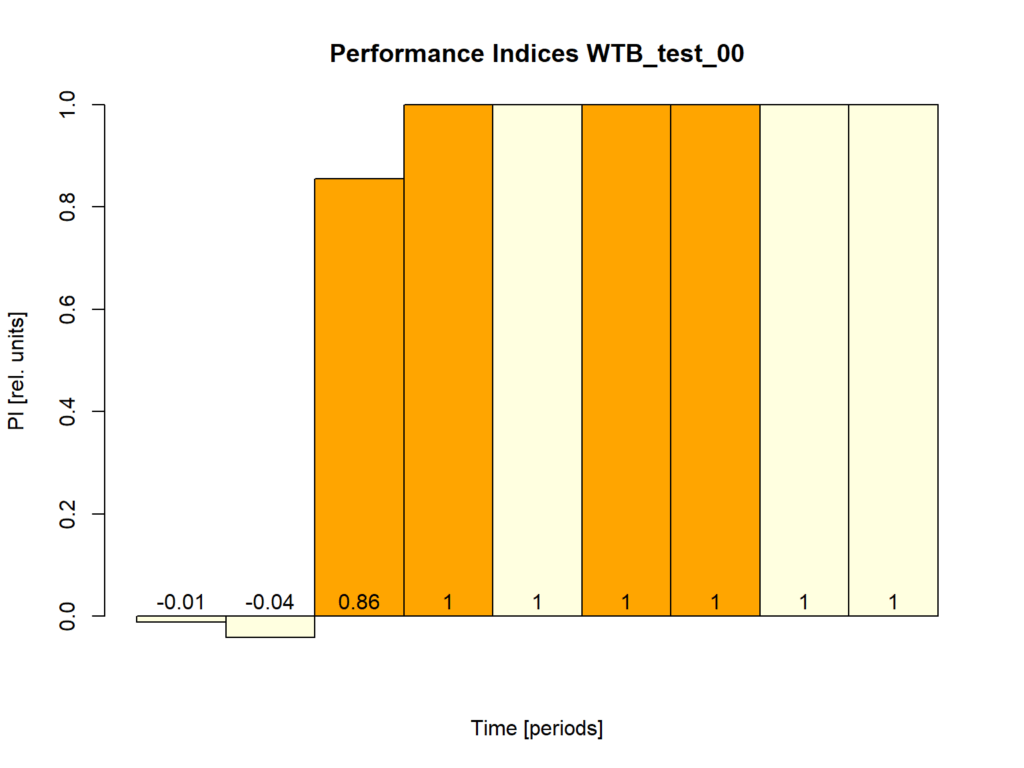
Category: Uncategorized | No Comments
Summary molecular work
on Monday, October 13th, 2025 1:58 | by Julia Schulz
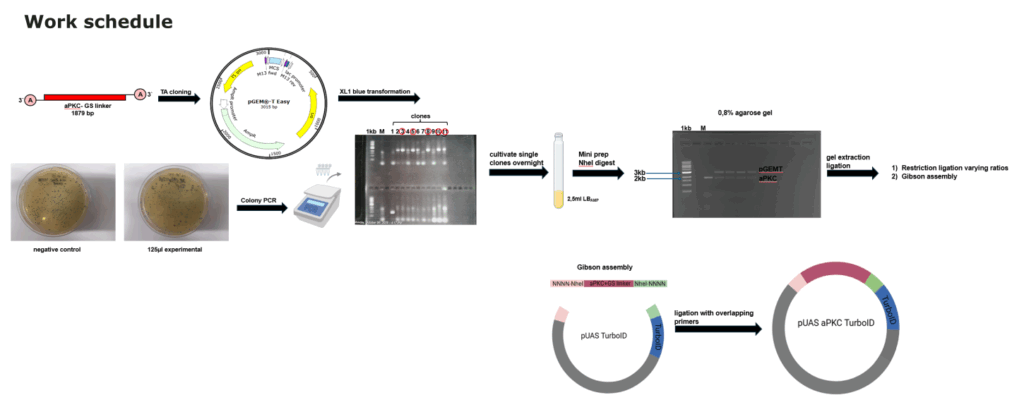
Category: Uncategorized | No Comments
Yt learning_positioning effect
on Monday, June 30th, 2025 11:47 | by Julia Schulz
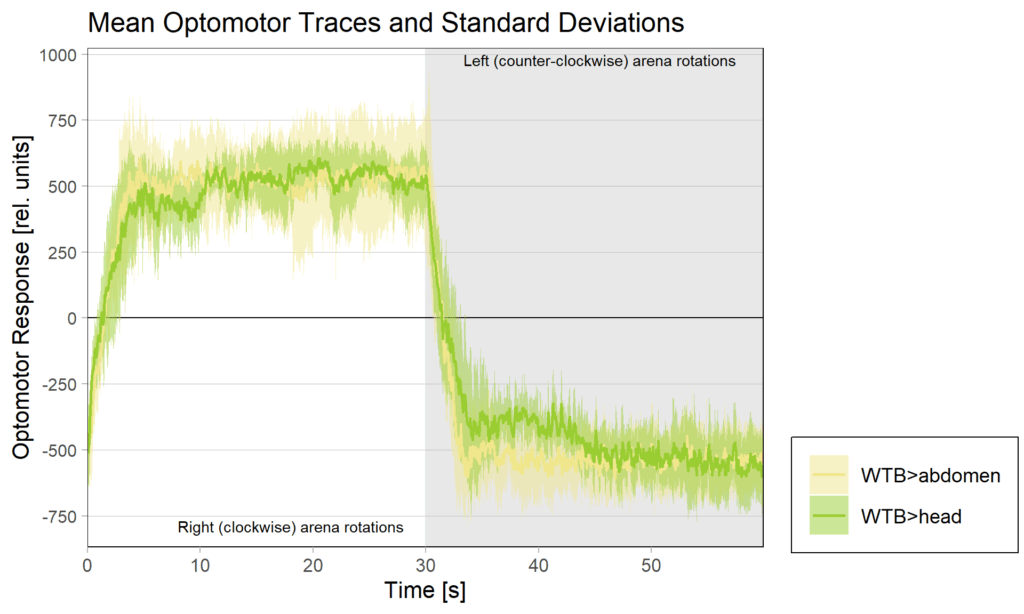
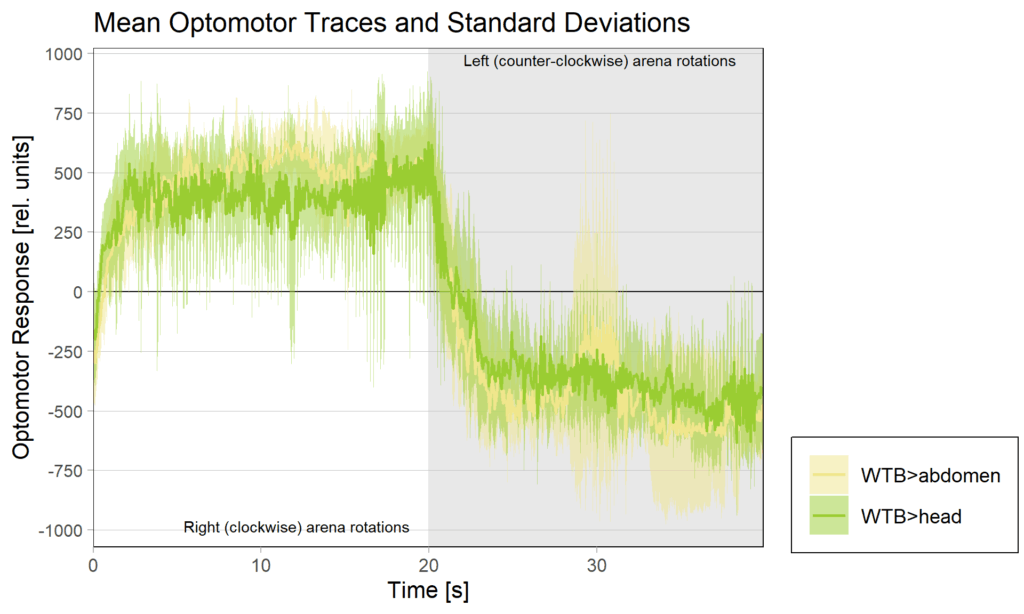
side bias can be compensated for by changing the positioning of the flies within the copper clamp from the medial to the lateral side facing away from the wall.
Category: Operant learning, operant self-learning, Uncategorized | No Comments
