Wild Type Drosophila memory ability test
on Monday, November 7th, 2016 11:07 | by Weitian Sun
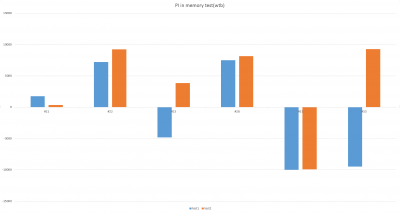
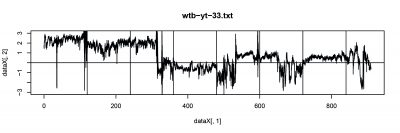
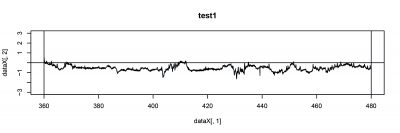
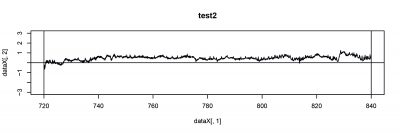
There were 6 more flies were tested, and PI from two tests are focused this time. The inconstancy in PI (#23, #33) from test1 and test2 indicates the learning or memory function in wild type during test.
- PI from all 6 groups
- fly trace(#33)
- test1 (#33)
- test2(#33)
Category: operant self-learning | No Comments
Wild type Drosophila flying trace
on Monday, October 31st, 2016 12:03 | by Weitian Sun
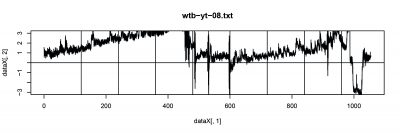
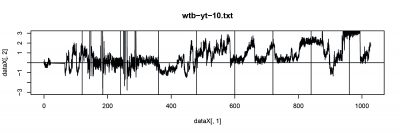
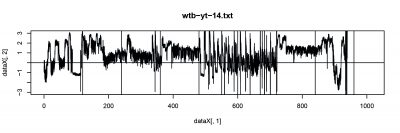
Wild Type Drosophila were used for single fly trace testing. The basic ideal is to record the spontaneous behavior (flying trace) after punishment training by heat. The followings are 3 subjects were test with performance index (PI).
- wtb_08 (right punishment).
| pretest(PI1) | training1(PI2) | training2(PI3) | test1(PI4) | training3(PI5) | traing4PI(PI6) | test2/3(PI7) |
| 10000 | 10000 | 10000 | 9984 | 9117 | 8113 | 10000 |
- wtb_10 (left punishment).
| pretest | training1 | training2 | test1 | training3 | traing4 | test2/3 |
| 1121 | 2583 | 2397 | -4310 | 9433 | 9458 | 9960 |
- wtb_14 (left punishment).
| pretest | training1 | training2 | test1 | training3 | traing4 | test2/3 |
| -832 | -9510 | -8339 | -9903 | -813 | -574 | -9887 |
- Average.
| pretest1/2 | training1 | training2 | test1 | training3 | traing4 | test2/3 | |
| all | 3429 | 1024 | 1352 | -1409 | 5912 | 5665 | 3357 |
| left punishment | 144 | -3463 | -2971 | -7106 | 4310 | 4442 | 36 |
PS: From Julien’s paper, he demonstrates that there are 7 blocks were included in PI ( one pretest:PI1; four training test: PI2, PI3, PI5,PI6; two memory test: PI4,PI7)
Category: Operant learning, operant self-learning | No Comments
UAS-PKC53eRNAI
on Friday, June 21st, 2013 6:47 | by Julien Colomb
So I got new flies to test. After one day of testing, I had only one control fly going through the test, all the 5 other were dead. That one did not learn.
I thus confirm that there is a problem with these flies. This means that I have no phenotype with one RNAi, and one phenotype with the other non-triggered RNAi which is probably not self-learning specific. This also means that the RNAi induction may not be sufficient and that I have no positive control for that.
The RNAi data are therefore not conclusive at all (neither positive nor negative results can be linked to any gene).
PS: I have checked the flies and overexpression of GFP did occur after 2 days Heatshock.
I will check the mutant again next week and I will be over with learning experiments. Still need some anatomy but then, I can finish the paper.
Category: Operant learning, operant self-learning | No Comments
not enough data to be sure- new cross needed
on Monday, May 13th, 2013 3:16 | by Julien Colomb
I will first test the mutant, if there is a phenotype, I will cross the RNAi lines to d42Gal4 to avoid using temperature shifts, which seems to be deteriorating the UAS RNAi vienna control line (only 4 flies made it to the test, lots of them died during the overnight heat shock).
Category: operant self-learning | No Comments
c380 driven PKC inhibition
on Tuesday, December 18th, 2012 5:25 | by Julien Colomb
Unfortunately, the amount of data is a bit too low to draw conclusion, but if one look at the data for each fly:
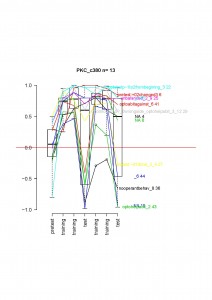
Only one fly not learning and the median would be 0. It looks very similar in the d42xPKCi cross, suggesting that the flies will also show the phenotype.
This will need to be confirmed next year with a new cross.
Other data visible at https://figshare.com/preview/_preview/104549
and later at doi: 10.6084/m9.figshare.104549
Category: operant self-learning, PKC, PKC_localisation | No Comments
c232,105yx UAS-TNT self learning
on Saturday, October 27th, 2012 4:44 | by Julien Colomb
none of the group learn. Flies are quite weak. The two groups with UAS show a clear problem in operant behavior (low score during learning phases)…
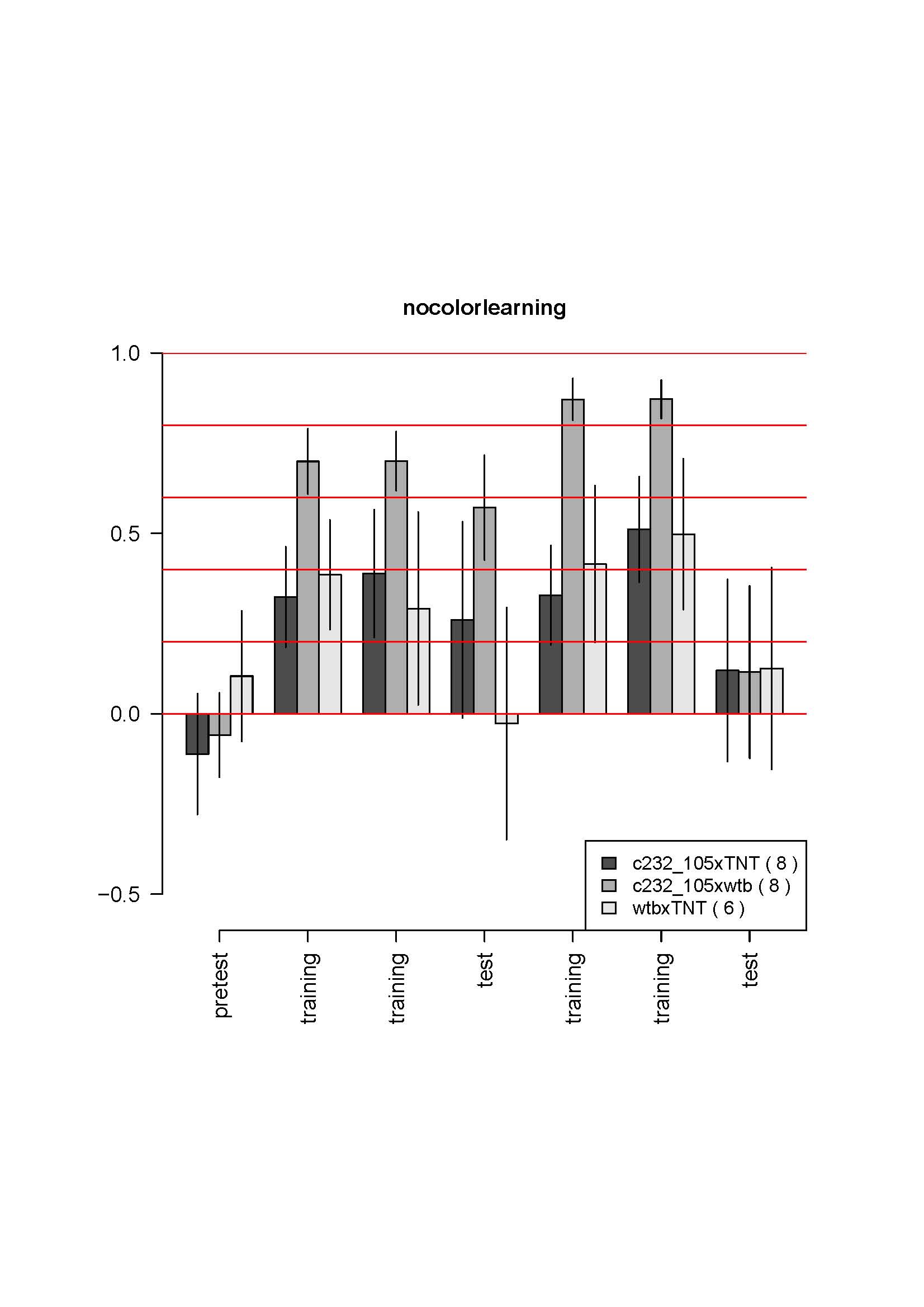
Category: operant self-learning, Spontaneous Behavior | 3 Comments
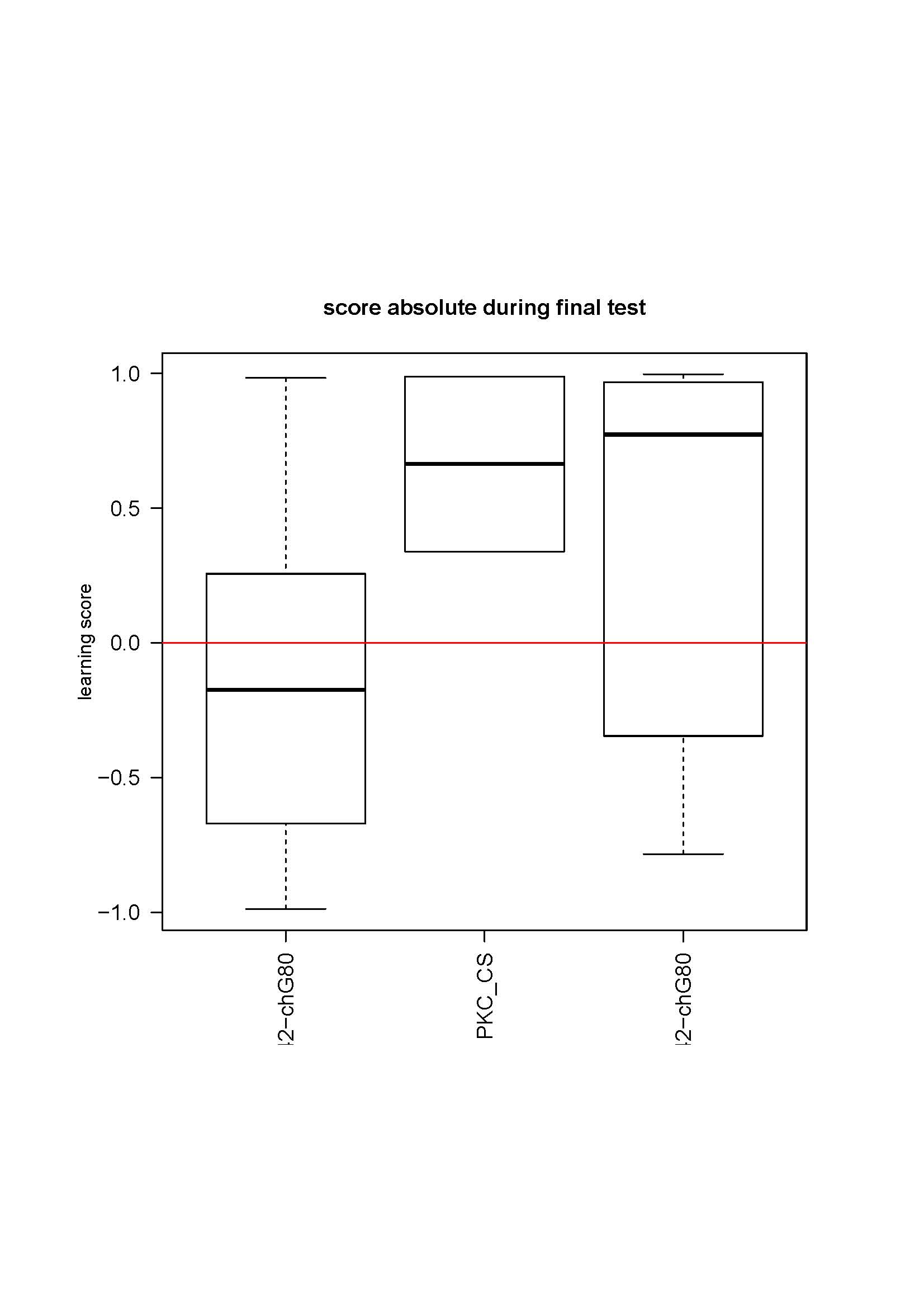
d42Gal4,chaGal80 (follow up)
on Wednesday, October 24th, 2012 3:05 | by Julien Colomb
Here is the data for first two crosses:
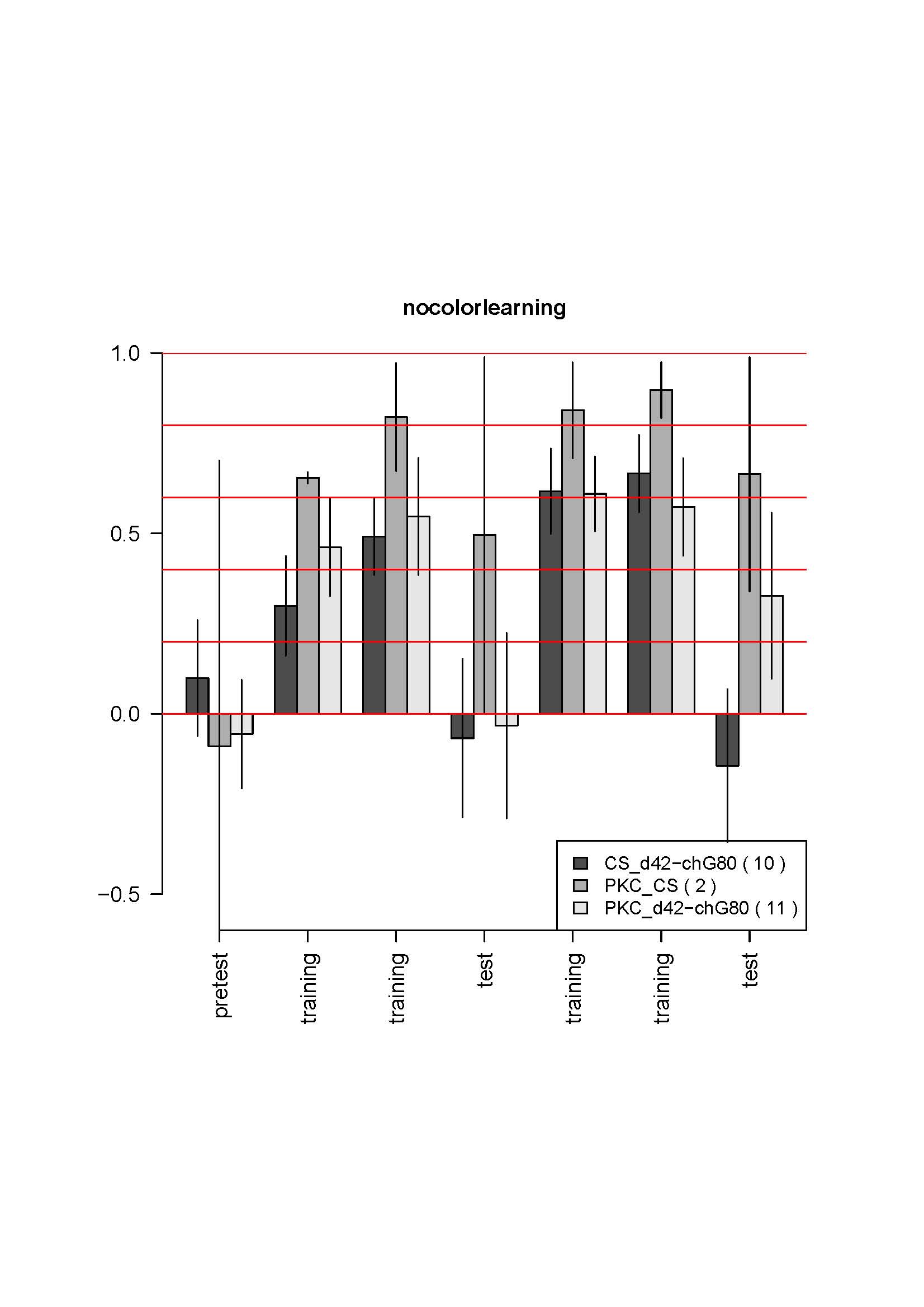
No changes from the first 4-6 tests. The most striking is the absence of learning for the control. It does not make sense. I also controlled that the genotypes were not inverted by looking at GFP fluorescence which is much weaker in the test line (due to the tubGal80ts presence) than in the control line. Could it be an effect of GFP presence in some neurons? The CS strain should be unguilty, since the CSx c380 had no phenotype in the previous cross…
A third cross is coming, with the elavGal4 control, maybe it is only bad luck… But the results were so reproductible so far, and so solid… it is strange. I got the parental lines out of the stock to get fresh flies for the next cross.
I should also cross new flies now. I am planning these groups (male-female):
UGFP;;d42gal4,chaGal80 x tubGal80ts,U-PKCi
CS x tubGal80ts,U-PKCi
UGFP;;d42gal4,chaGal80 x CS
UGFP,c380;;,chaGal80 x tubGal80ts,U-PKCi
Category: operant self-learning, PKC | 1 Comment
SMap before and after self-learning: no difference
on Friday, October 19th, 2012 6:33 | by Julien Colomb
small but significantly different from 0 slope in the S-Map procedure, both before and after self-learning.
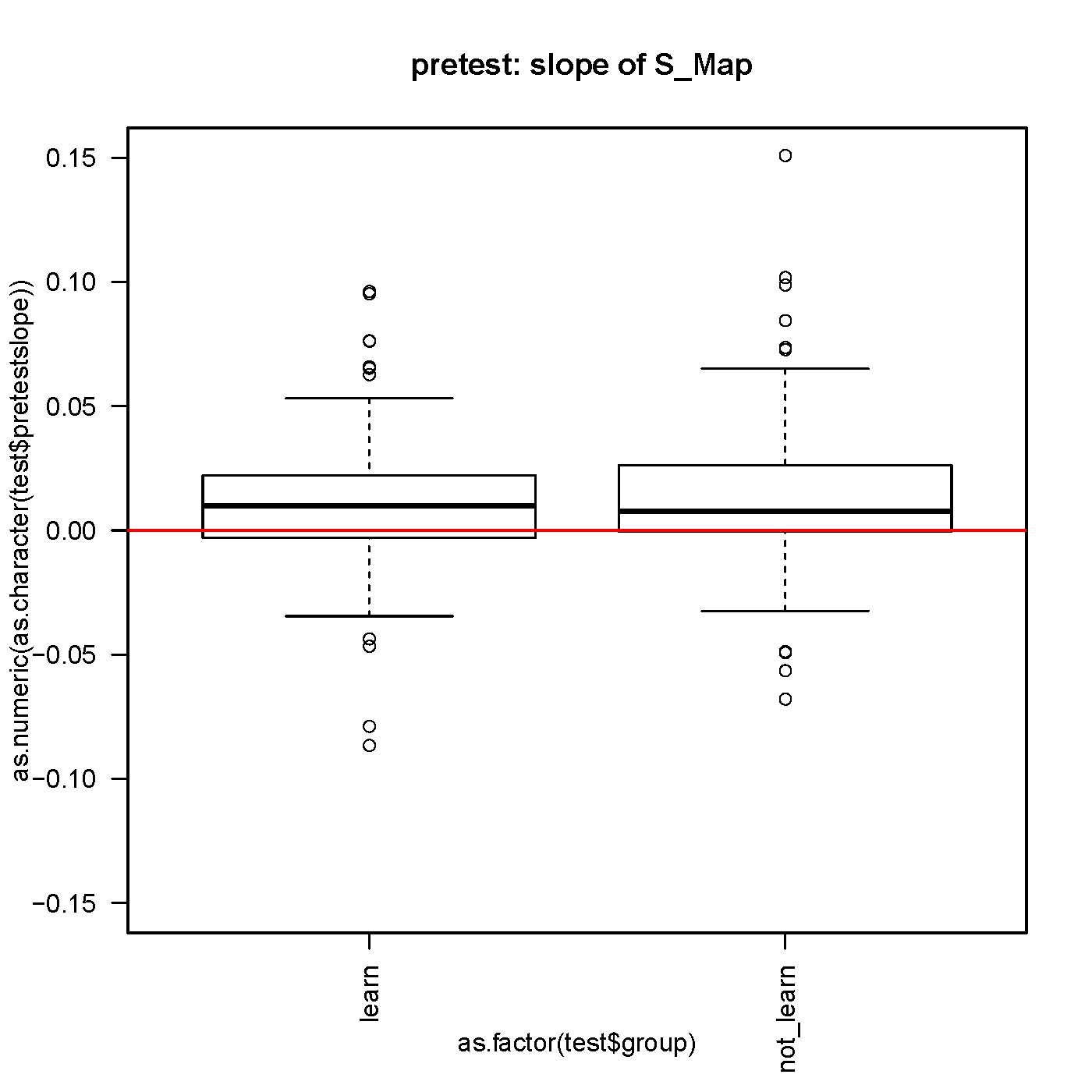
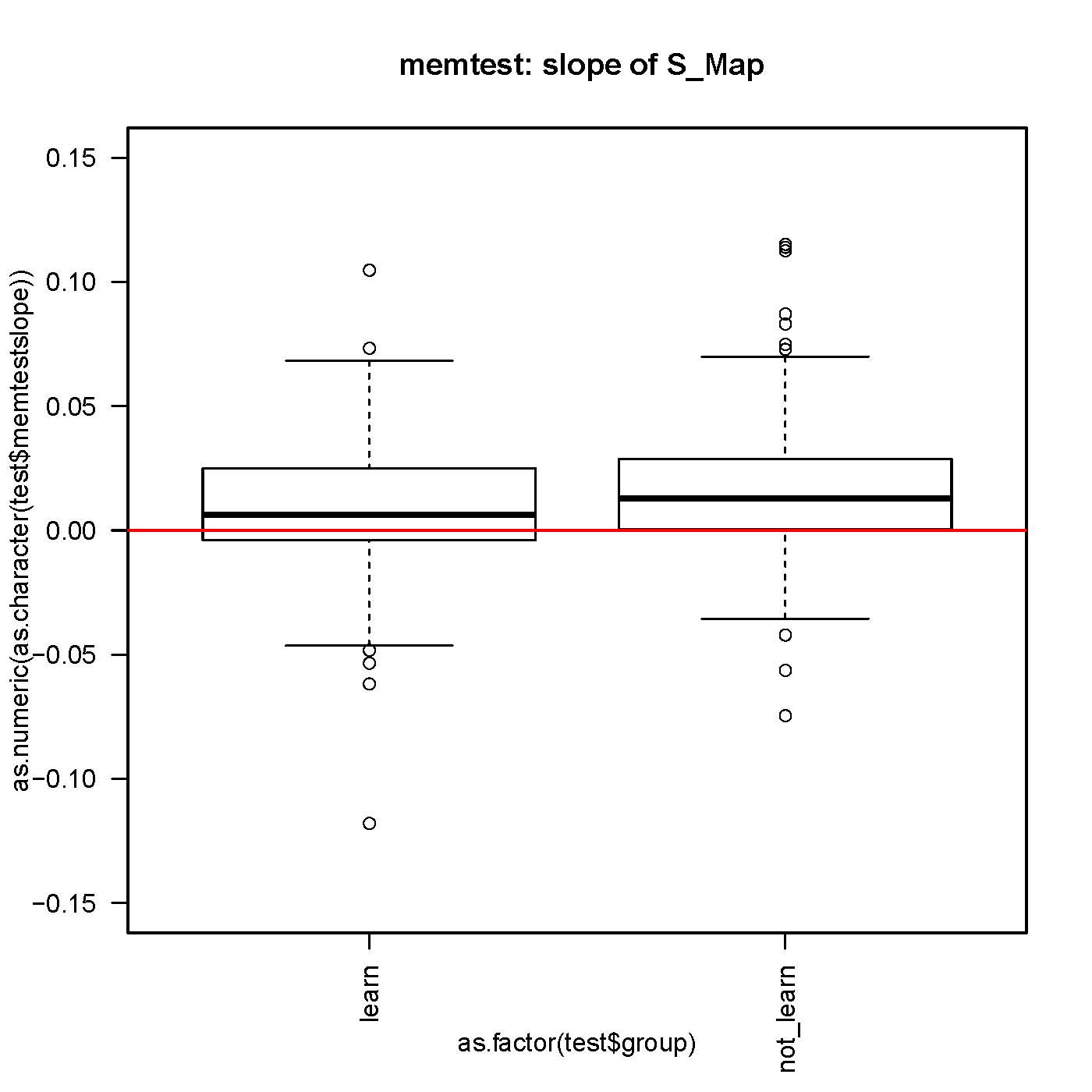
No difference in the slope while comparing before and after learning for each fly.
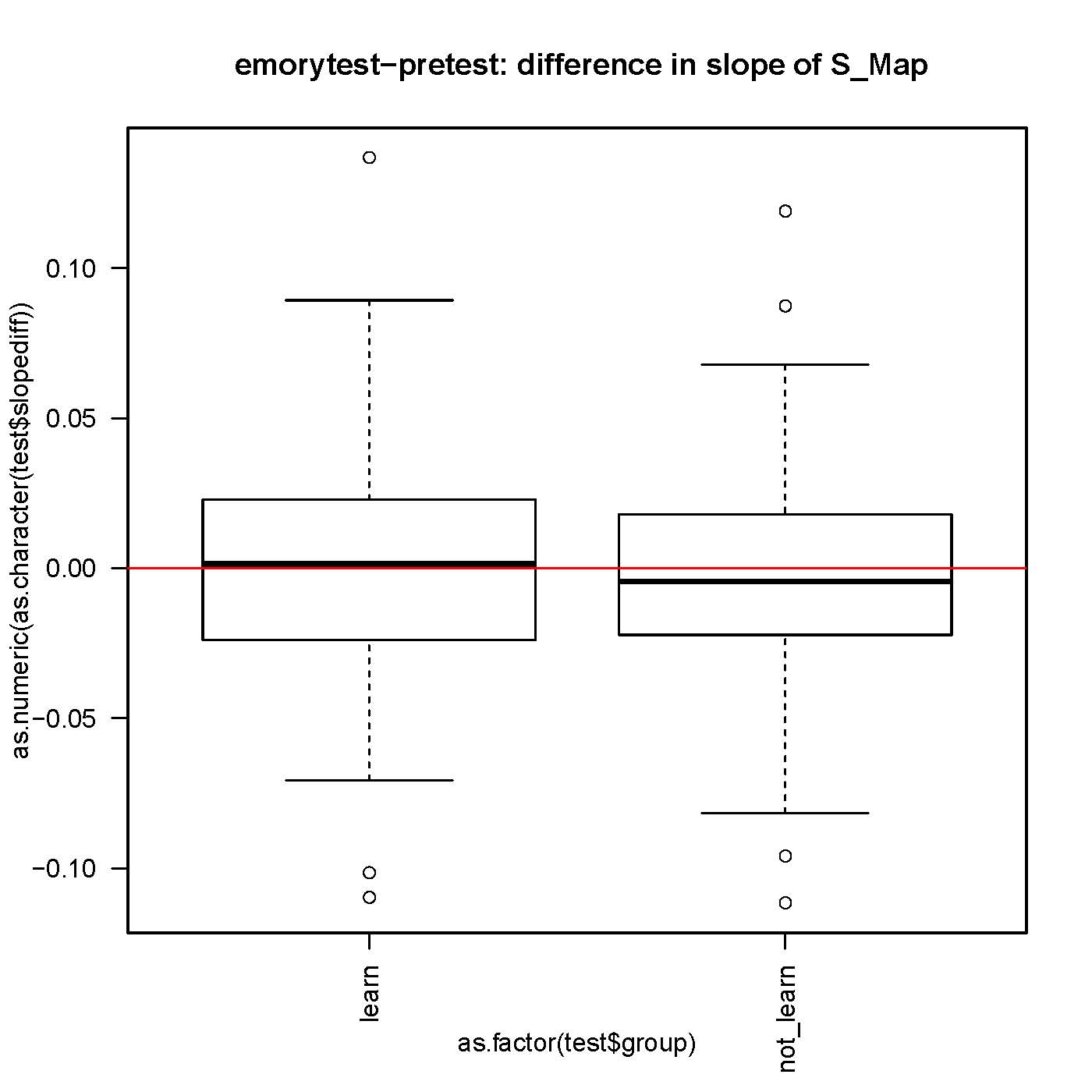
n>130 for each group.
Category: operant self-learning, Spontaneous Behavior, Uncategorized | 4 Comments
FoxP anatomy
on Thursday, October 18th, 2012 5:05 | by Julien Colomb
I had a look at the brain preparation I did last week (dicer2,FoxP RNAi x elavGal4 and controls, with nc82 antibody immunostaining). They look surprisingly good under the fluorescent microscope.
We still need to make confocal scans. I will ask Hilde whether she would be interested to do it…
Category: Foxp, operant self-learning | No Comments
PKC53E
on Thursday, October 18th, 2012 2:58 | by Julien Colomb
I got mutant from Amita and will test them when out of quarantine. Then, we will have a clear answer about the compensatory mechanisms (the former mutant I tested was probably not null). There is also 2 other RNAi lines for PKC53e from the TriP project.
I am waiting for chrisi to be back and we will see what flies we need to order to bloomington in addition to these two. (There is also another RNAi against FoxP…)
The outcross of the RNAi is also running (3d cross, now).
We will then have 3 RNAi and one mutant, if the results are consistent, this would be enough to convince people that PKC53e is the PKC involved.
Category: operant self-learning, PKC | 2 Comments