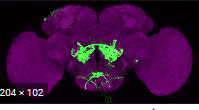
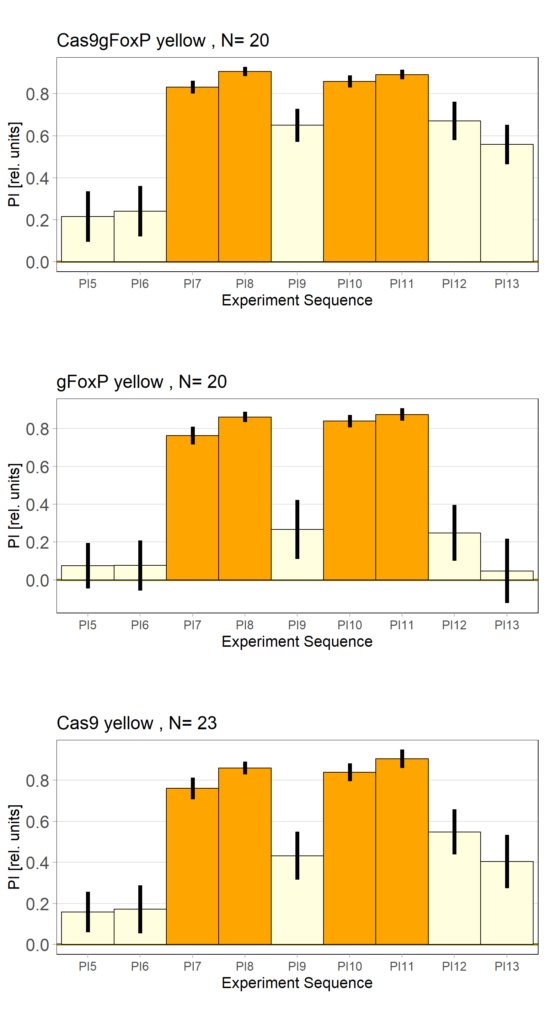
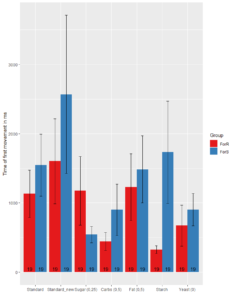
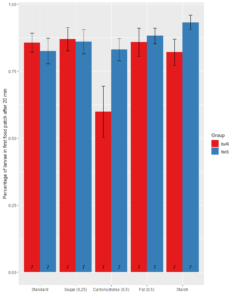
Local FoxP knok out GMR11F02-GAL4
on Monday, February 15th, 2021 9:30 | by Andreas Ehweiner
Category: flight, Foxp, Memory, Operant learning, operant self-learning, Uncategorized | No Comments
Larval food manipulation evaluation
on Monday, February 8th, 2021 1:28 | by Simon Benmaamar
1. Food Patch
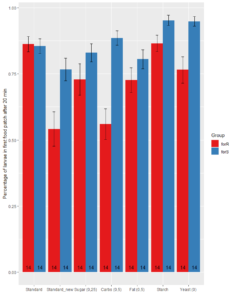
Significant difference between Rover and Sitter in Standard_new (wilcox.test, p=0.01057), Carbohydrates (wilcox.test, p= 0.0002445), Starch (wilcox.test, p-value = 0.03431) and Yeast (wilcox.test, p = 0.001455)
2. Distance Tracking
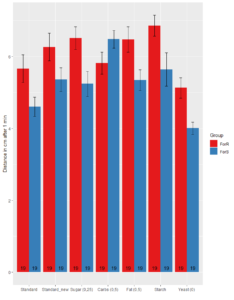
No significant difference between Rover and Sitter in Standard_new (t-test, p =0.08108), Carbohydrates (t-test, p=0.09875),
Category: Rover/Sitter, Uncategorized | No Comments
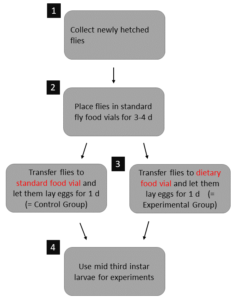
Larval food manipulation first evaluation
on Monday, February 1st, 2021 12:40 | by Simon Benmaamar
Food Patch Experiment
Category: Uncategorized | No Comments
Optogenetics- Joystick (THD’, THD1, Gr28bd-TrpA1)
on Monday, February 1st, 2021 11:39 | by Aida Kumpf
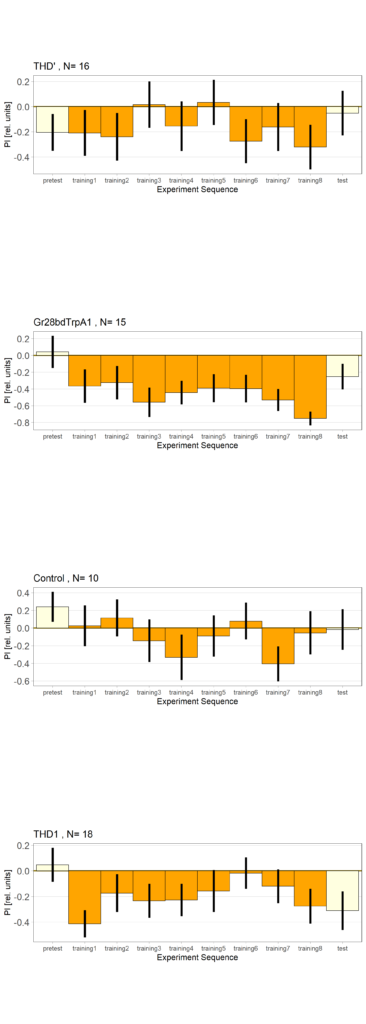
Category: Uncategorized | No Comments
Walking speed and stripe deviation: Buridan’s paradigm
on Monday, January 25th, 2021 1:01 | by Sayani Banerjee
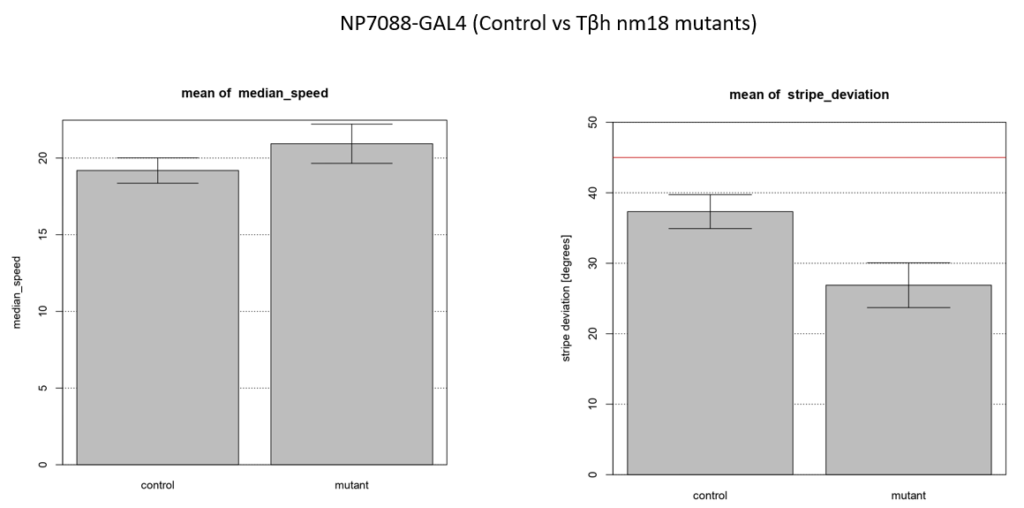
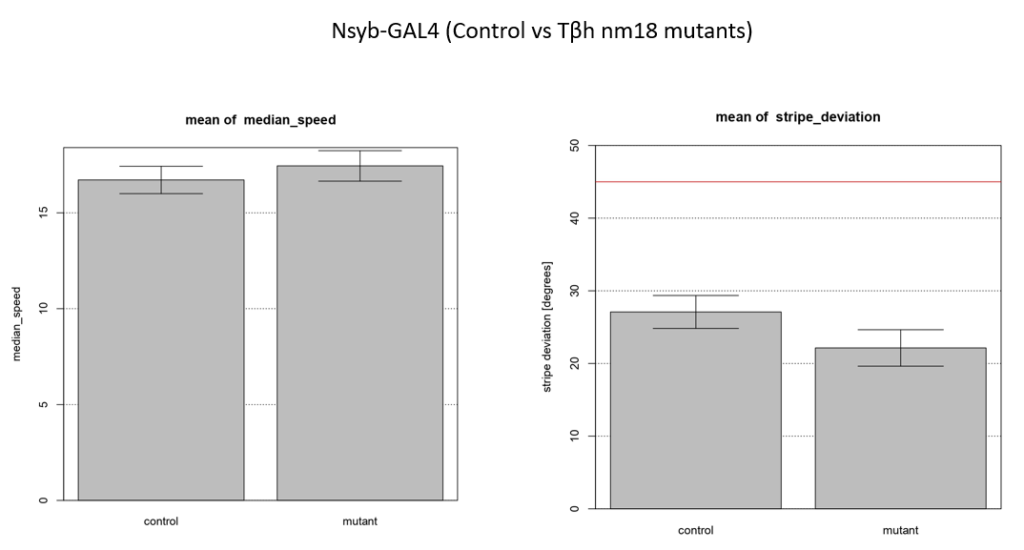
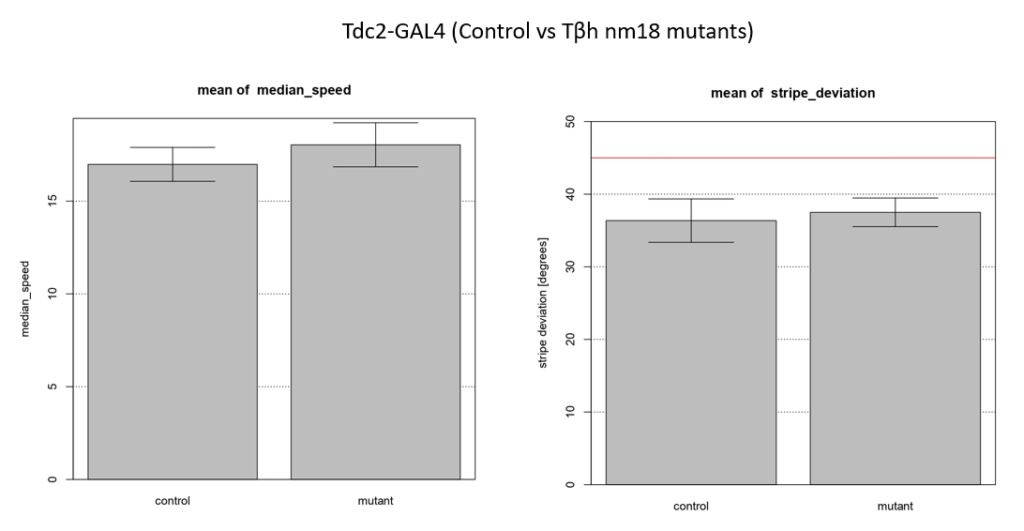
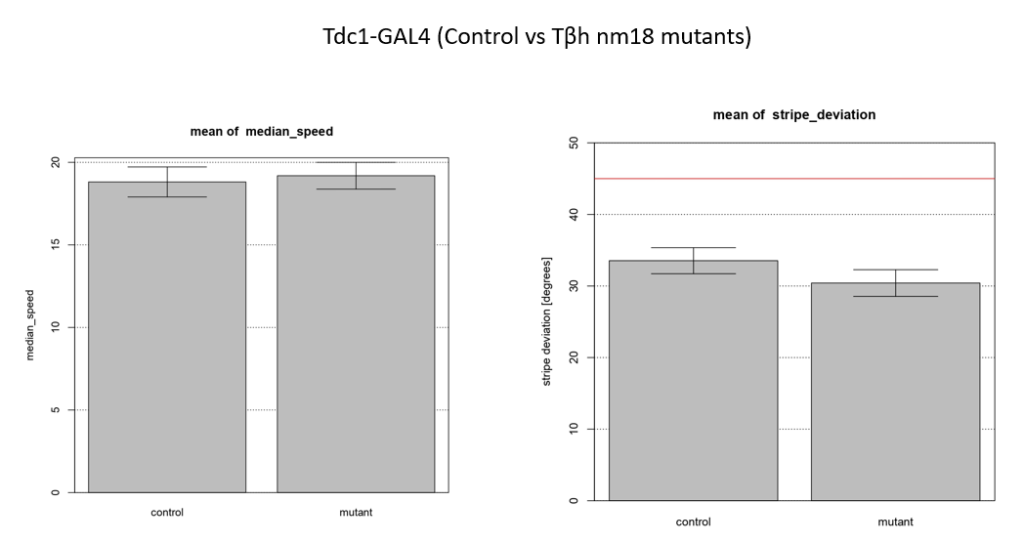
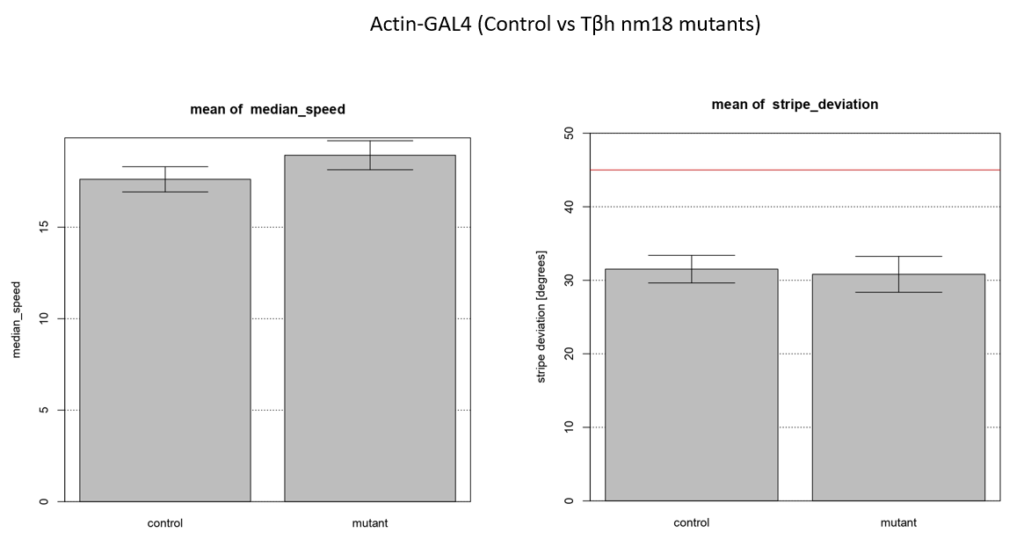
Category: Uncategorized | No Comments
To compare with previous results
on Monday, January 11th, 2021 1:17 | by Sayani Banerjee
Control vs Tbhnm18 mutant ( -GAL4)
Category: Uncategorized | No Comments
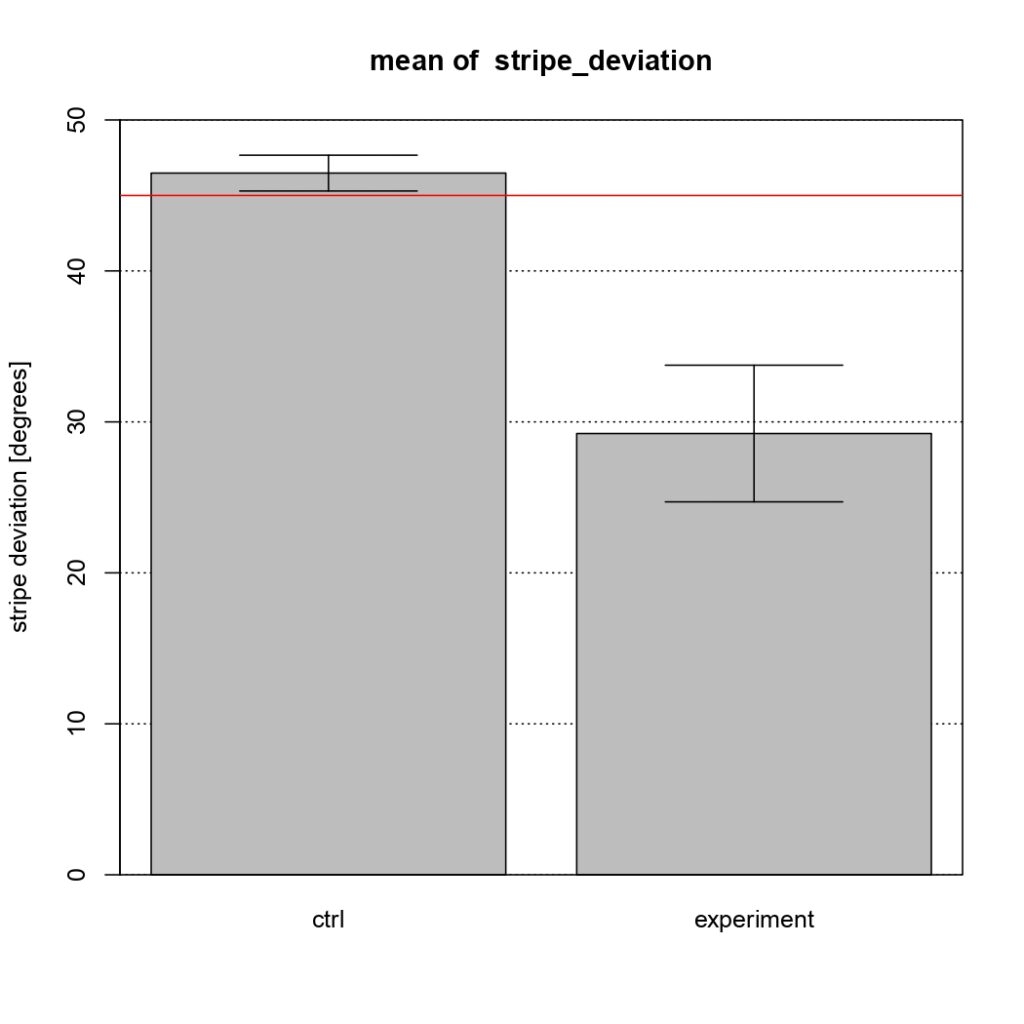
NSyb-GAL4, mean of stripe deviation
on Monday, January 11th, 2021 12:58 | by Sayani Banerjee
Control vs Tbhnm18 mutant
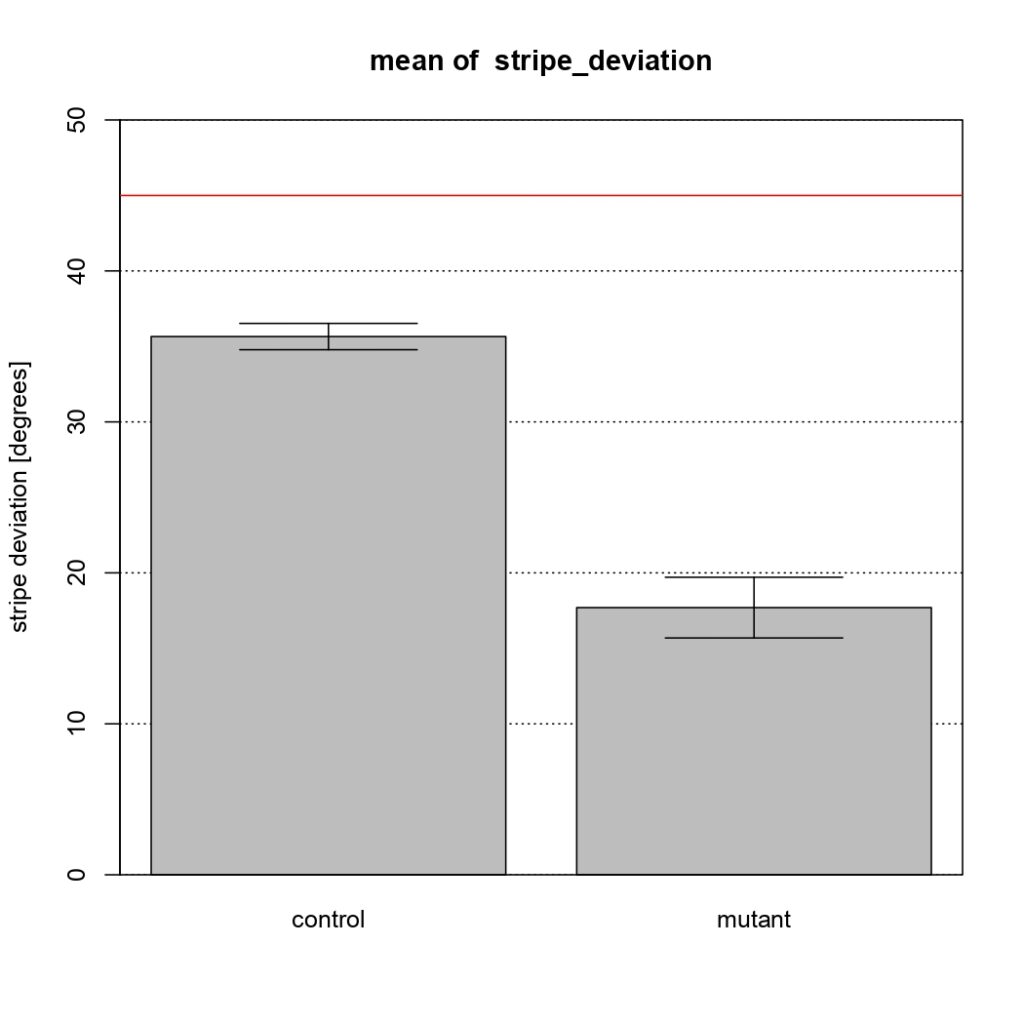
Category: Uncategorized | No Comments
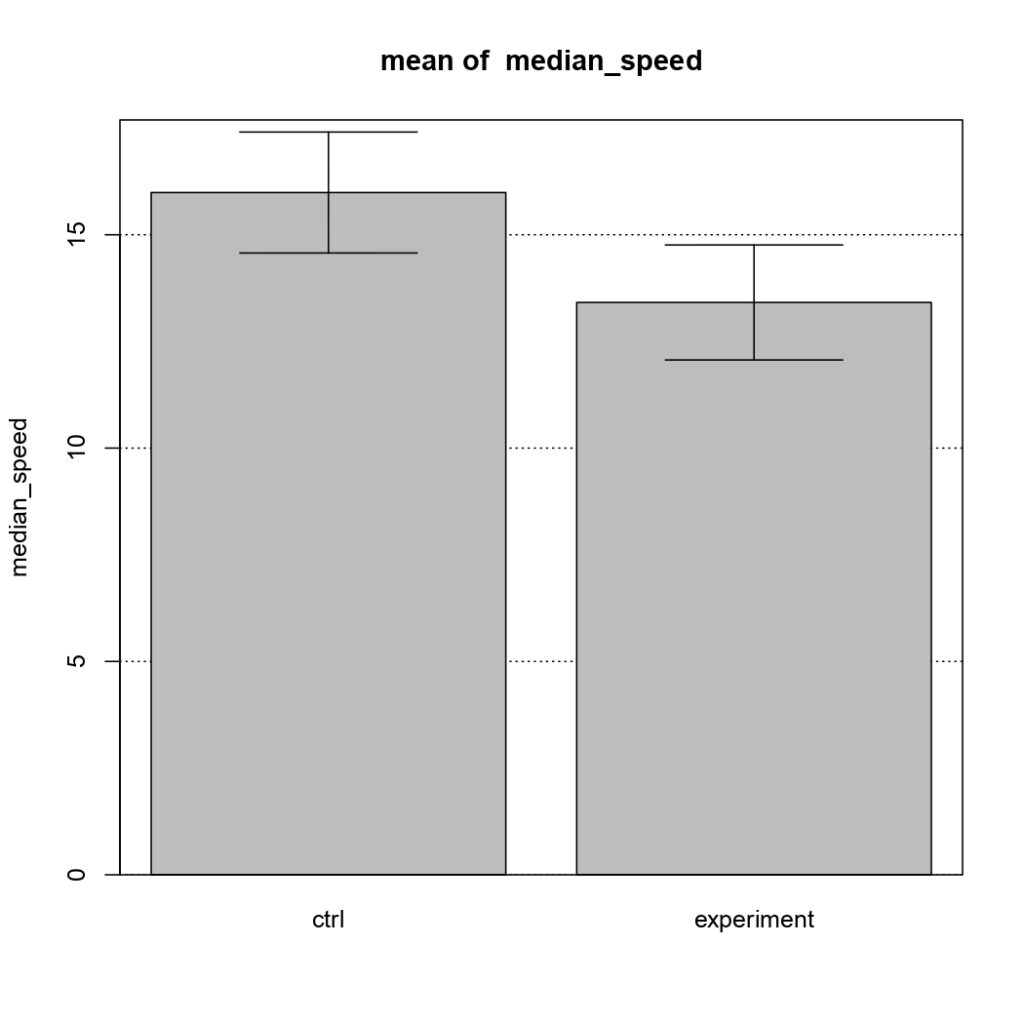
NSyb-GAL4, mean of median speed
on Monday, January 11th, 2021 12:56 | by Sayani Banerjee
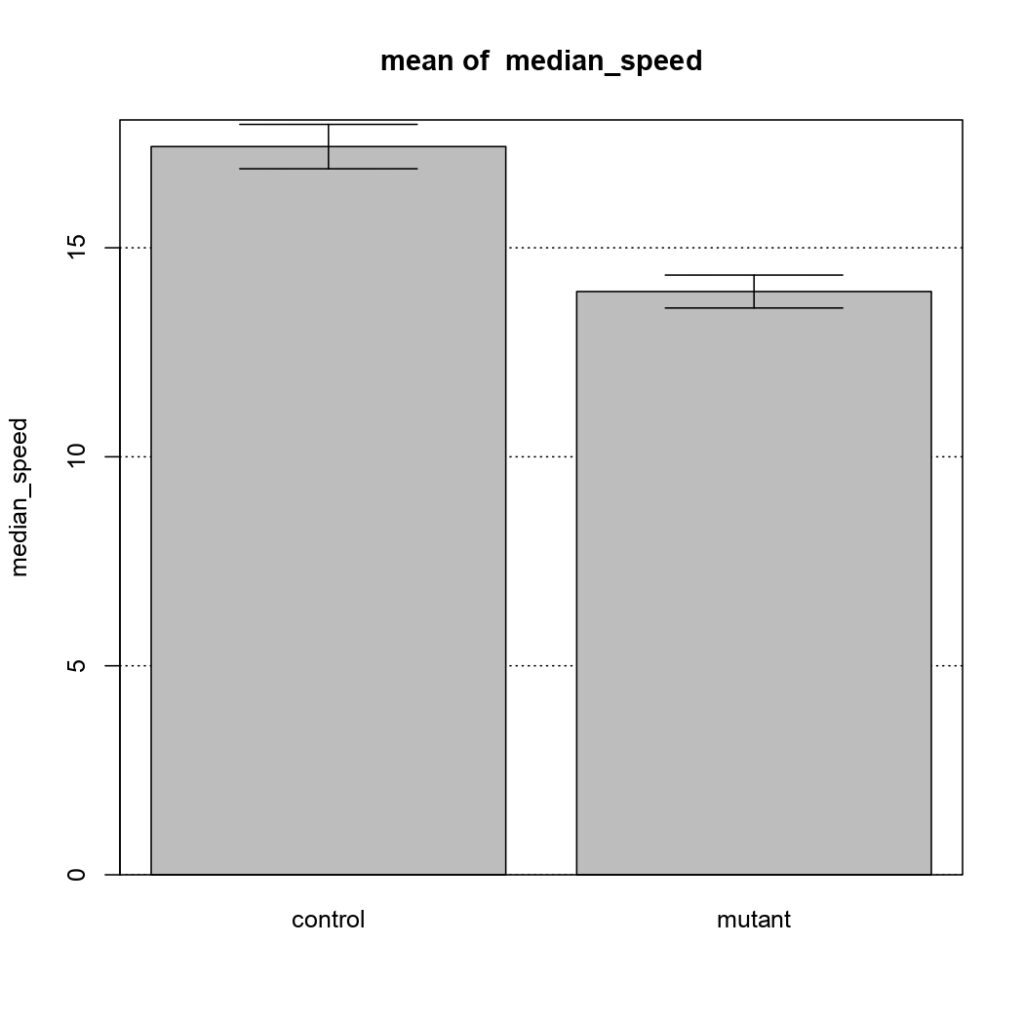
Category: Uncategorized | No Comments
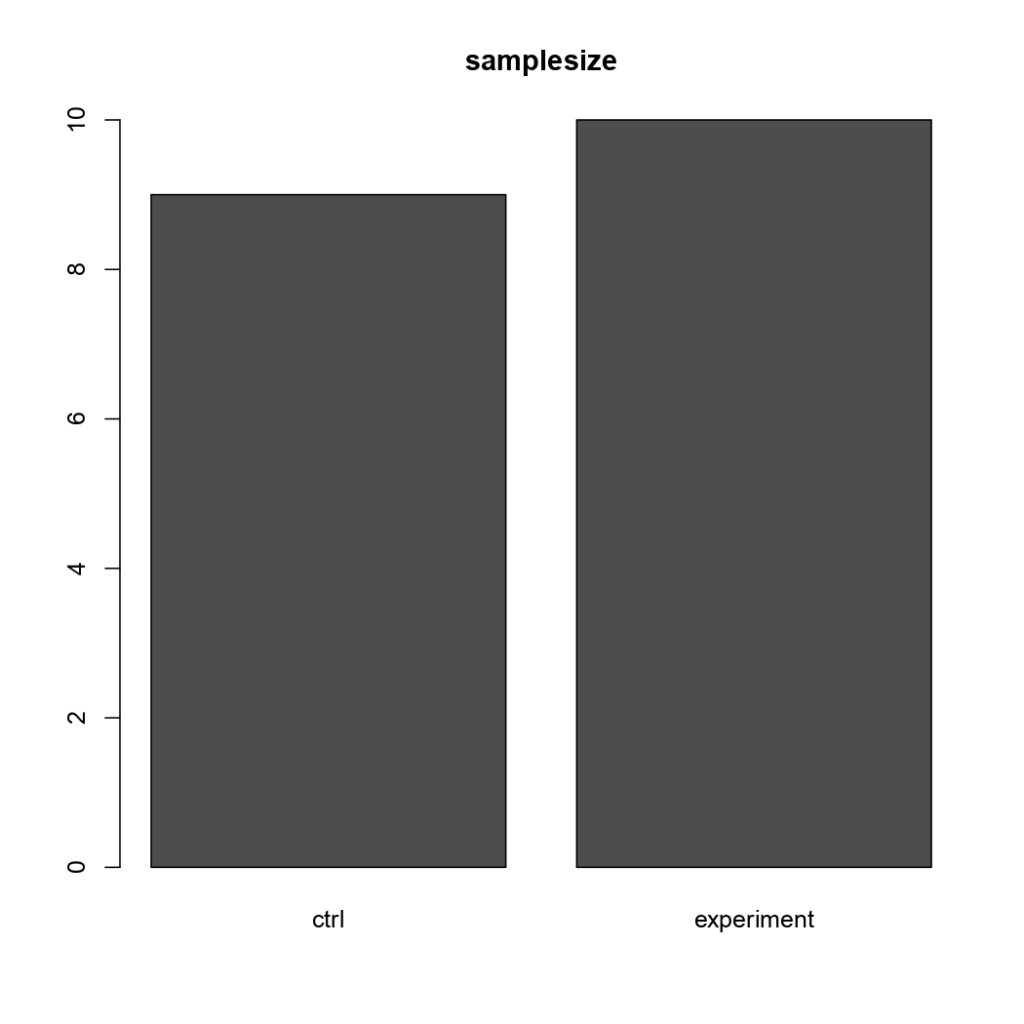
NSyb-GAL4, Sample Size
on Monday, January 11th, 2021 12:52 | by Sayani Banerjee
(Control vs Tbhnm18 mutant)
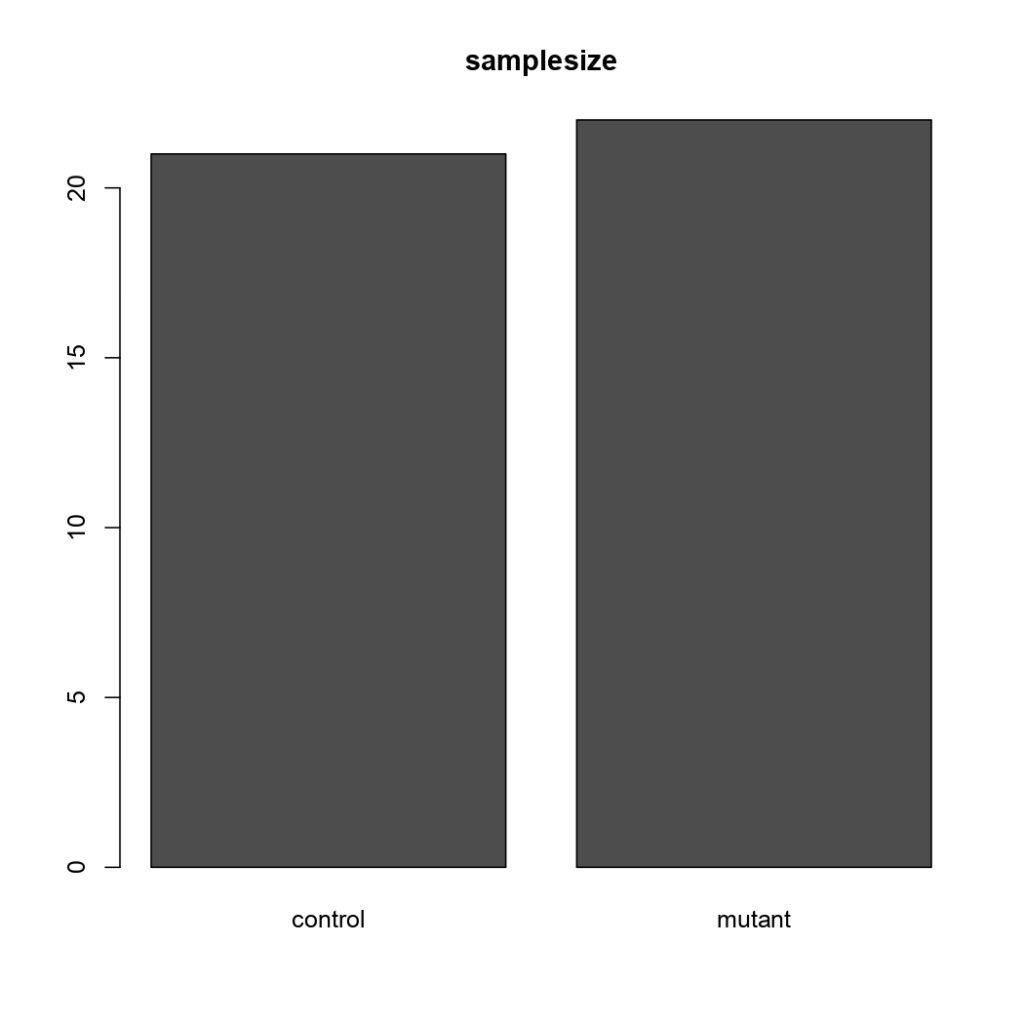
Category: Uncategorized | No Comments
Planned experiments
on Monday, December 21st, 2020 11:37 | by Simon Benmaamar
Category: Rover/Sitter, Uncategorized | No Comments