LiLiu data on buridan
on Thursday, November 8th, 2012 2:50 | by Julien Colomb
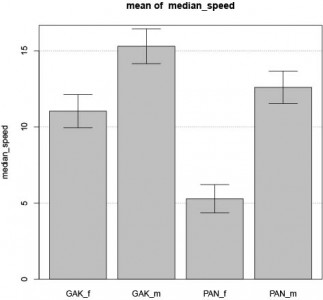
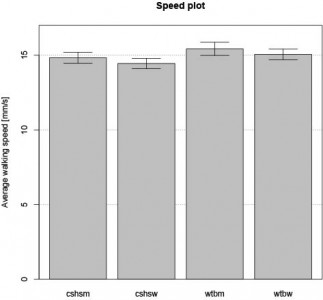
I started to work with the data I got from Li Liu. I have no metadata file, so I am not sure what will finally come out of the analysis.
What is already clear is that the number of female and male they tested seem different for each group and there is a big difference in speed between male and female… The value reported are also huge (>20 mm/s, no distinction male/female) while I get the normal 15mm/s (if the data I got is in mm and not in pixel, of course).
I do not know what GAK and ‘PAN stands for…
Remember that from our data, there is no significant difference in male versus female speed.
Category: buridan, open science | No Comments
First flight tests, Trehalose again…
on Thursday, November 1st, 2012 7:33 | by Christine Damrau
Chrisi’s day…
1 Collected virgins for our ill post-doc, made some crosses:
d42-GAL4 males x tubGAL80-UAS-PKCi and x CS
d42-chaGAL80 males x tubGAL80-UAS-PKCi and x CS
2 Buridan: Prepared colored food (10mg/ml, 2.5ml total in the very little vials, ~ 5 flies in each vial). Denise tested in the afternoon females in the Buridan. Data (4n now) will be shown in the lab meeting.
3 Imaging Made a new cross for the Imaging-project:
TDC2-GAL4 males x UAS-GCaMP3(2)
replaced Neloy’s flies by flies from stock
Florian’s larval preparation (good!) showed that GCaMP-expression is much less than GFP-expression. They hope for the homozygous line.
4 Flight: Performed flight experiments with the tbh,TDC,UAStbh flies. Raw data, see below. Thinking about analysis (probably tomorrow)
Protocol: glued the day before, sugar in wheel chambers, little EVIAN-water.
5s filter on the legs before starting, aspirated, stopped when no response 3 times in a row, stopped time when 30min, stopped after 15 aspiration trials
| date | fly | genotype | number_aspiration | time_to_first_stop | duration_of_flights | remarks |
| 01.11.2012 | 1 | 12 | 4.56 | 4.56,0.22,0.30,0.2 | ||
| 01.11.2012 | 2 | 2 | 49.15 | 49.15,14.42 | ||
| 01.11.2012 | 3 | 16 | 0.03 | 0.03,0.15,3.06,1.56,0.3,0.14,4.58,0.22 | ||
| 01.11.2012 | 4 | 3 | 0 | 0 | ||
| 01.11.2012 | 5 | 40 | 0.05 | 0.05,0.01,0.02,0.01,0.01,0.02,0.02,0.01,0.02,0.03,0.02,0.02,0.01,0.01,0.01,0.01,0.01,0.04,0.09,0.01,0.02,0.01,0.11,0.01,0.01,0.01,0.01,0.01,0.02,0.01,0.03 | ||
| 01.11.2012 | 6 | 3 | 0 | |||
| 01.11.2012 | 7 | 13 | 3.01 | 0.01,0.01,0.01,0.01,0.01,0.02,0.01 | ||
| 01.11.2012 | 8 | 15 | 0.01 | 0.01,0.04,0.03,0.02,1.56,0.15,0.02,1.46,3.26,1.15,0.46,6.45,2.56,0.06,12.59 | ||
| 01.11.2012 | 9 | 15 | 0.01 | 0.01,0.01,0.28,0.1,1.5,1.29,0.40,0.29,1.34,0.54,0.22,0.37,0.31,0.54 | ||
| 01.11.2012 | 10 | 9 | 0.01 | 0.01,0.01,0.01,0.01,0.01 | ||
| 01.11.2012 | 11 | 12 | 0.01 | 0.01,0.01,0.01,0.01,0.01,0.01 | ||
| 01.11.2012 | 12 | 3 | 0 | |||
| 01.11.2012 | 13 | 4 | 0.06 | 0.06,0.5,0.06,38.2 | ||
| 01.11.2012 | 14 | 3 | 0.19 | 0.19,0.33,0.31,30 | ||
| 01.11.2012 | 15 | 15 | 3.38 | 3.38,2.43,0.47,0.36,4.06,0.39,1.25,0.35,0.52,1.07,0.22,0.23,0.27 | ||
| 01.11.2012 | 16 | 3 | 0 | |||
| 01.11.2012 | 17 | 3 | 7.16 | 7.16,23.27,30 | ||
| 01.11.2012 | 18 | 3 | 0 | |||
| 01.11.2012 | 19 | 6 | 1.33 | 1.33,14.57,9.17,30 | ||
| 01.11.2012 | 20 | 1 | 18.20 | stopped the expriment | ||
| 01.11.2012 | 21 | 3 | 2.50 | 2.50,2.18 | stopped the expriment |
5 Trehalose: Found time to try out Gérard’s idea (no pigment in the probes to not disturb the NanoDrop measurement). 4 groups: 10 flies with or without heads, 50 flies with or without heads. Hilde showed me how to remove heads with liquid nitrogen and vortex. The other flies (with heads) were frozen at -20° as usual. Trehalase incubation over night. Tomorrow glucose oxidase and measurement
Category: Biogenic Amines, buridan, flight, trehalose measurement | No Comments
First lab book post
on Wednesday, October 31st, 2012 6:47 | by Christine Damrau
Hi all, I forgot about the lab book email, because it reached me in New Orleans… My day:
1 Collected virgins for our ill post-doc
2 Prepared colored food (10mg/ml, 2.5ml total in the very little vials, ~ 7 flies in each vial). Vicky tested in the afternoon females in the Buridan. The the duration of drug application was increased to 3h because there was no effect after 1 hour. Data will be shown in the lab meeting.
3 Answered to Dennis Pauls to make an appointment for the Trehalose measurement in Würzburg
4 Glued hooks for a flight experiment I did not do because I did not know, how… (idea was to test whether the UAS-tbh cross is ok)
5 Checked my TDCxGCaMP flies for expression in the fluorescence dissection scope (very cool!) in Marco’s room (borrowed from Konstantin). Then realized that there are pupa in the virgin vials :(
6 Temperature-dependent experiments in Buridan: wanted to test the infrared lamp with the dimmer (measure the temperature change). Was impossible because the thermometer itself heated up. So, I finally looked for an infrared thermometer (you can point on the area you want to measure).
7 boring stuff… Dienstreiseabrechnung was wrong… Email to the Chinese guys again about the cylinder for Buridan… Explained Florian (Marco’s master student) some fly stuff…
Category: Biogenic Amines, buridan | No Comments
sathish’ log
on Wednesday, October 24th, 2012 11:43 | by Sathish K Raja
1. Working on the buridan speed threshold activity data.
2. I mailed to Dr.Thomas Binz who supplied TNT antobody to Heisenberg lab and awaiting for response.
3. C105-Gal4 will be out of quarantine within this week.
4. Working on the thesis writing, gathering information about tetanus toxin expression studies done by sweeney.
Category: buridan | No Comments
GRIP on Buridan conti.,
on Friday, October 19th, 2012 7:17 | by Sathish K Raja
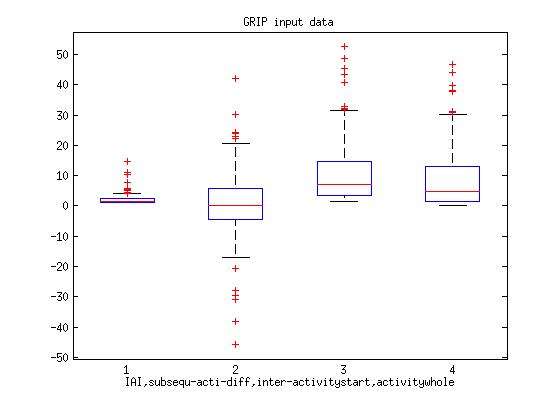
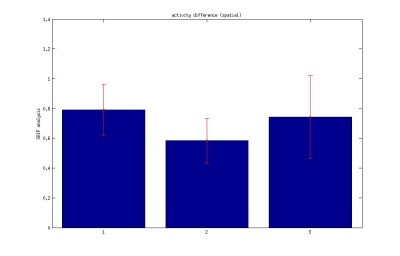
Here I post the figures belong to,
I. Boxplot of one fly input data to GRIP.
Legend: 1.Time interval between each activity (Pause length)
2.Difference between subsequent activity (subtracting the subsequent activity)
3. Times between start of activity
4. Time duration of each Activity (activity data itself)
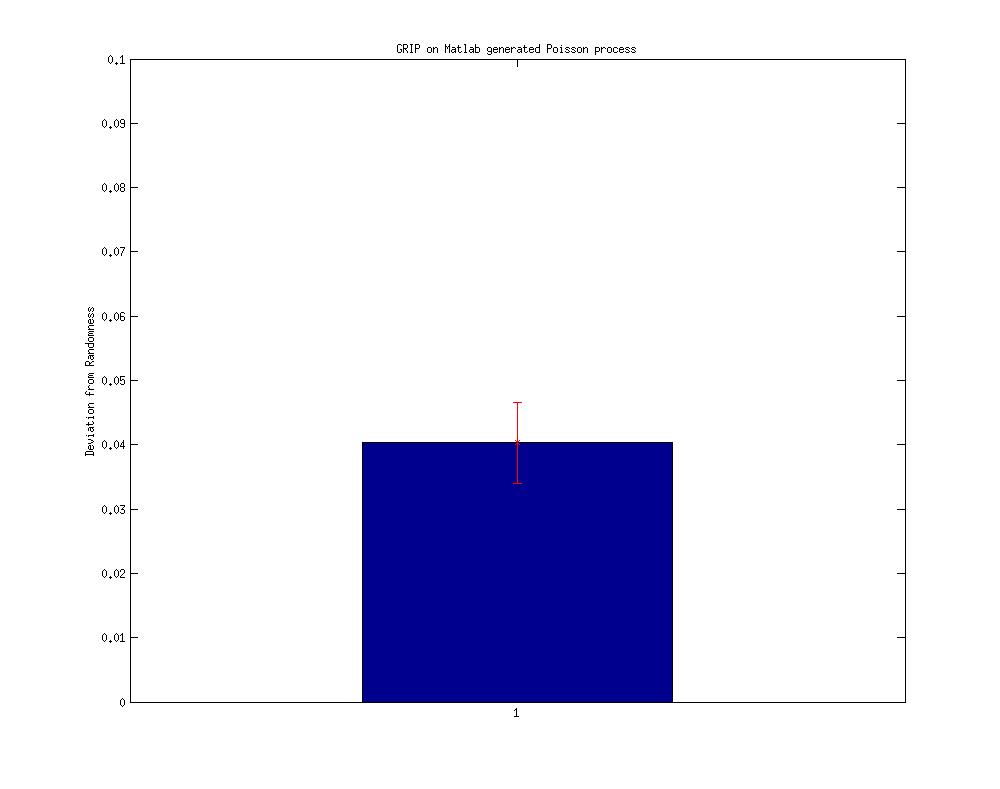
II. GRIP on Matlab generated pseudo-Poisson process (on the modified code )
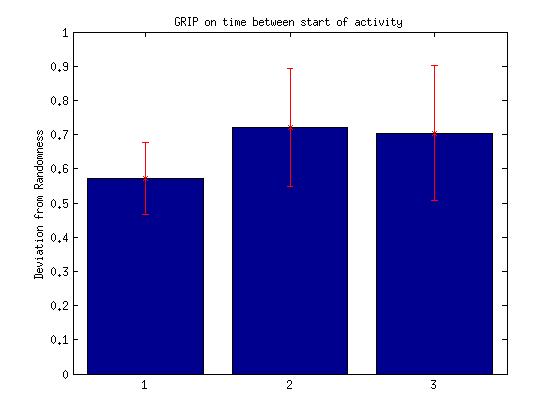
III.Times between animal starts
Bar: 1.c105;c232-wtb
2.c105232-TNT
3. Wtb-TNT
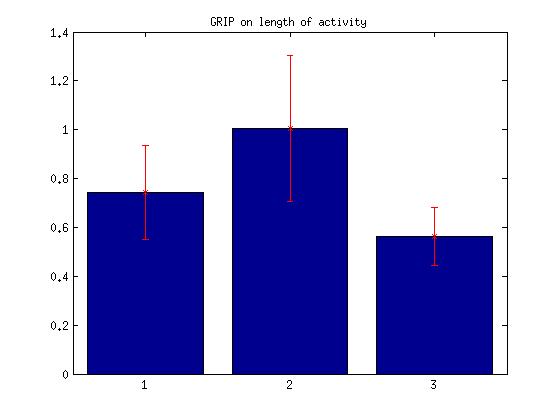
IV. Length of activity
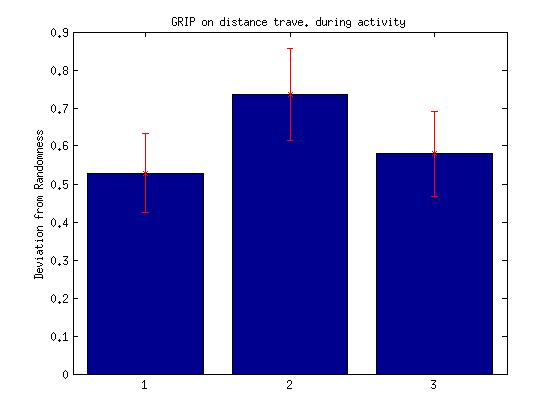
V. Total distance travelled during activity period
Category: buridan | 3 Comments
grip analysis on Buridan activity data
on Wednesday, October 17th, 2012 7:49 | by Sathish K Raja
Grip analysis on:
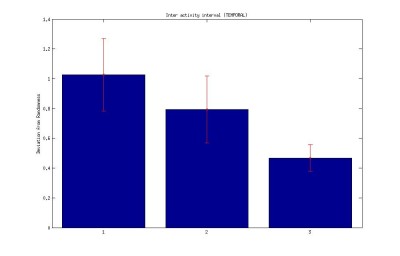
1. Interval between activities (temporal)
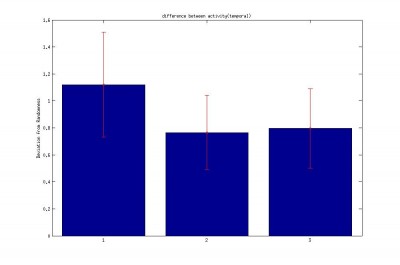
2.Difference between the subsequent activity (temporal) like [X2-X1,X3-X2]
3.Difference between the subsequent distance travelled during the activity period (spatial)
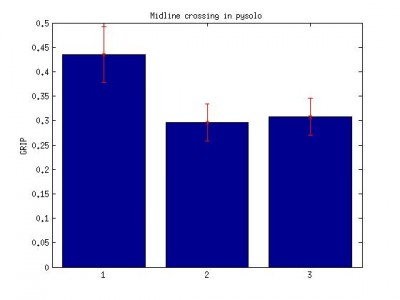
4.Pysolo midline crossing data. Data sampling at 1 min interval. number of time fly crosses the virtual midline over the period of 60 s.
Bar: 1.c105;c232-wtb
2.c105232-TNT
3. Wtb-TNT
Inter-activity interval of buridan activity
2.Difference between the subsequent activity
Difference between the subsequent distance travelled during the activity period
pysolo midlines crossing
Category: buridan, Pysolo, Spontaneous Behavior | 5 Comments
on Tuesday, October 16th, 2012 6:02 | by Julien Colomb
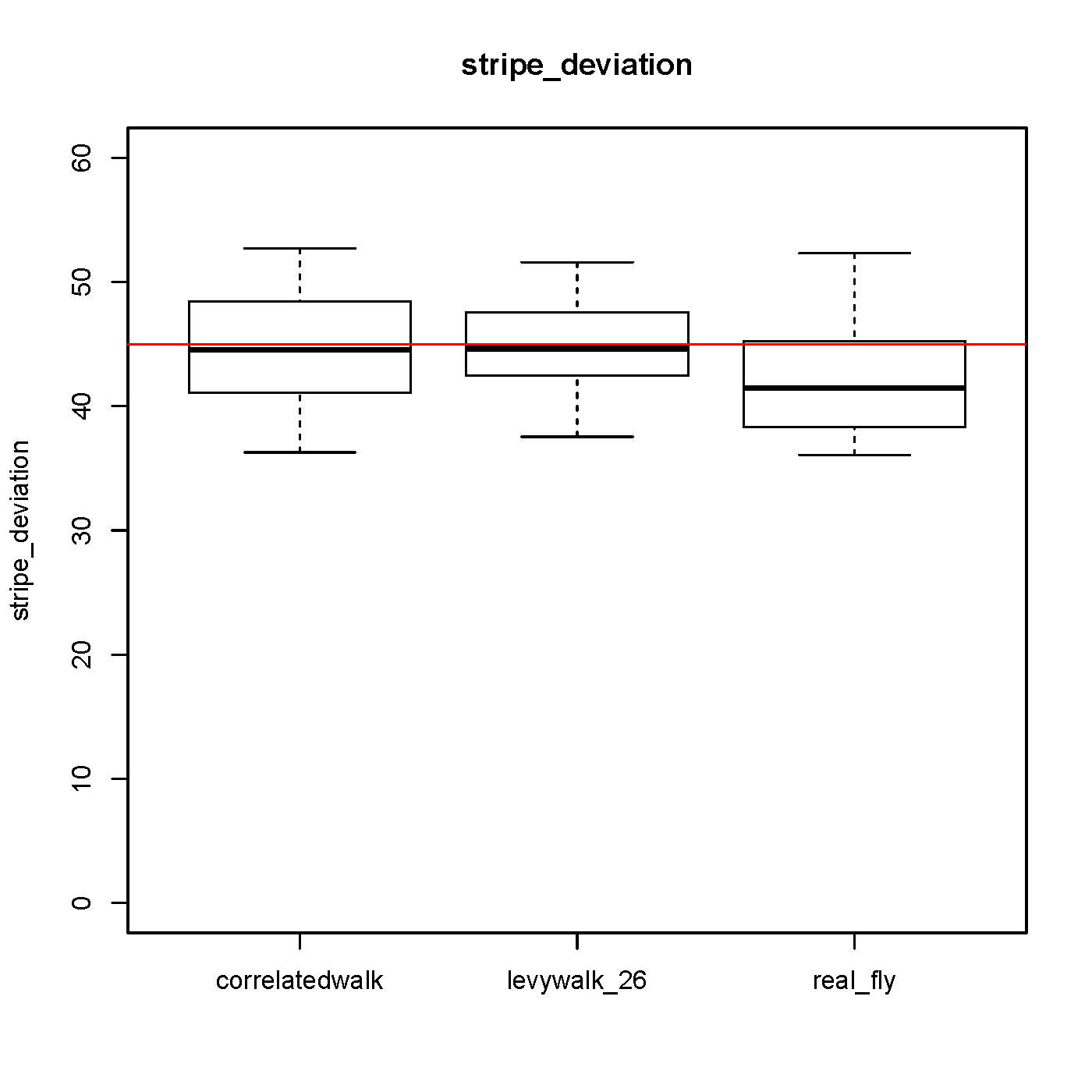
I worked on CeTrAn, corrected some bugs and added boxplot visualization of each variable. As an examples see how the stripe deviation variable vary in computer generated data, quite impressive:
Category: buridan, open science | No Comments
15 Oct Sathish’ log
on Monday, October 15th, 2012 6:43 | by Sathish K Raja
Well,
Since its lab notebook, I write in such a way that I could easily understand the tasks and rest could follow up. I will post figures with explanation and results whenever available.
Matlab:
1. working on the scripts to differentiate initial 2 min and last 2 min of self learning exp. to see the learning effect.
dataset suffix numbered in random fashion, unable to create handle to embed in struct field. Cell array conversion was possible but cant identify the fly specificities. Waiting to get updated version of data from Julien.
21 Hz corrected to 20 hz using interp1 method written and saved in home/character/Julien/
rest of code compiled and saved in same loc.
2. Buridan data – after analysis .csv file data is arranged on basis of activity and non activity.
wrote codes to extract each fly data out of .csv and place in .mat file to enable easy loop over. error due to presence of 2 activity log subsequently due to bursts inside activity matrix. troubled files isolated.
interval between each activity is considered as IAI . Even number logic is used in script. data(m,n)*ones(M)
sprintf(‘sol%d’,’iteration’) leaves only string ,convert using str2num and call the metafile.Yet to compile.
3.Altered the Grip code such that grip does not try to convert the location of activity(spikes) to time domains.saved as griburidan.m only used with buridan and pysolo data.yet to compile to function
Flies:
1.crosses under way for self learning with crowd control
2.c819 Gal4 arrived from Bloomington n quarantined.
Log end
Category: buridan, Spontaneous Behavior | No Comments
News from Neurofly
on Tuesday, September 11th, 2012 6:50 | by Julien Colomb
A lot of things I learned at the neurofly. First my old friend Benjamin is doing a great postdoc in Bruno van Swinderen lab. It was great to see his data on isoflurane effects and the correlation between sleep disorder and sensitivity to anesthesia.
More related to our interests, I learned that Paul Tchénio has now a prototype to visualize neuronal activity in about half of the fly brain at a decent frequency. It may be interesting to get in touch with him again.
Andre Fiala is doing/has already done the TDC-Flp construct and is using his own UAS-stop-TRPA1 line to get a subset of octopaminergic neurons. Christine will contact him soon to have more information and maybe start a collaboration with him. The student was not at his/her poster when we went there, and we could not talk directly to him/her.
I nice talk by F. Mohammad (Singapore) showed that centrophobism can be associated with anxiety: the majority of treatments used with mices (for instance immobilisation in a pipet tip) also induce more (or less for some treatments) centrophobisms in flies. I told him about the CeTrAn, he said he will look at it. (his flies were in a squared box, with their wings untouched).
Jhl21 (receptor for JH) change of larval behavior at the wandering stage (go slower and turn more): it acts at the NMJ to change the clustering of glutamate receptors. Similar thing may happen during the first hour of life of the fly, when they become phototactic ?
A nice poster from the Heisenberg’s lab show that flies do have a preferred 0 torque. Even when giving some closed loop drift, the histogram of torque suggest that the preference for the 0 torque is still there (non-uniform distribution around the +1 torque which stabilize the drift. It seems the distribution around the 0 torque is seen only if you have long enough data, I told them I would ask Satish to look at his distributions. This emphasizes why we need to be very careful in preparing the fly for our experiments…
I may have to look at the OK6 Gal4 line for motorneurons, and new RNAi lines seem to be available for PKC53e and FoxP…
I also talked to Flybase people, David was there. It seems the Buridan results should lead to 2 different entries: one linking each genotype to a dichotomic description of the phenotype (“mutant1” – “has larger”- “median_speed”) and one to the raw data and analysis. This will not be easy to automatize, but looks interesting.
I am probably forgetting a couple of things in addition to my discussion with Anette Schenk and her student Anna about FoxP..
buridan analysis v3.0
on Friday, August 19th, 2011 7:58 | by Julien Colomb
The new version of the analysis software is online. It should be clean and will be the version coming with the paper.
Category: buridan, Uncategorized | 1 Comment