JoyStick results with 650 Lux yellow light
on Monday, June 30th, 2025 12:08 | by Daniel Döringer
We were having some issues with our Negative Control for the JoyStick. Without ATR the flies should not avoid optogenetic activation since the channel should not be functional, yet still they did. We lowered the light intensity from 800 Lux to 650 to see whether the light was to strong and would activate the Chrimson channel, even without ATR.
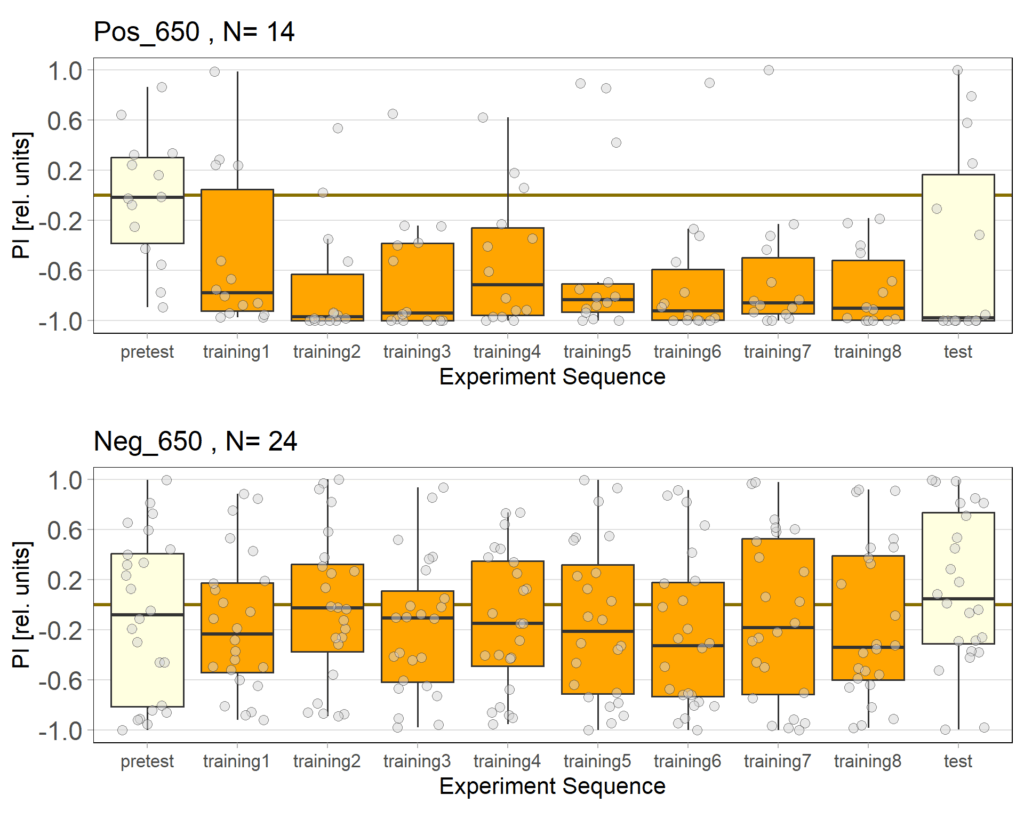
Alisa’s experiments
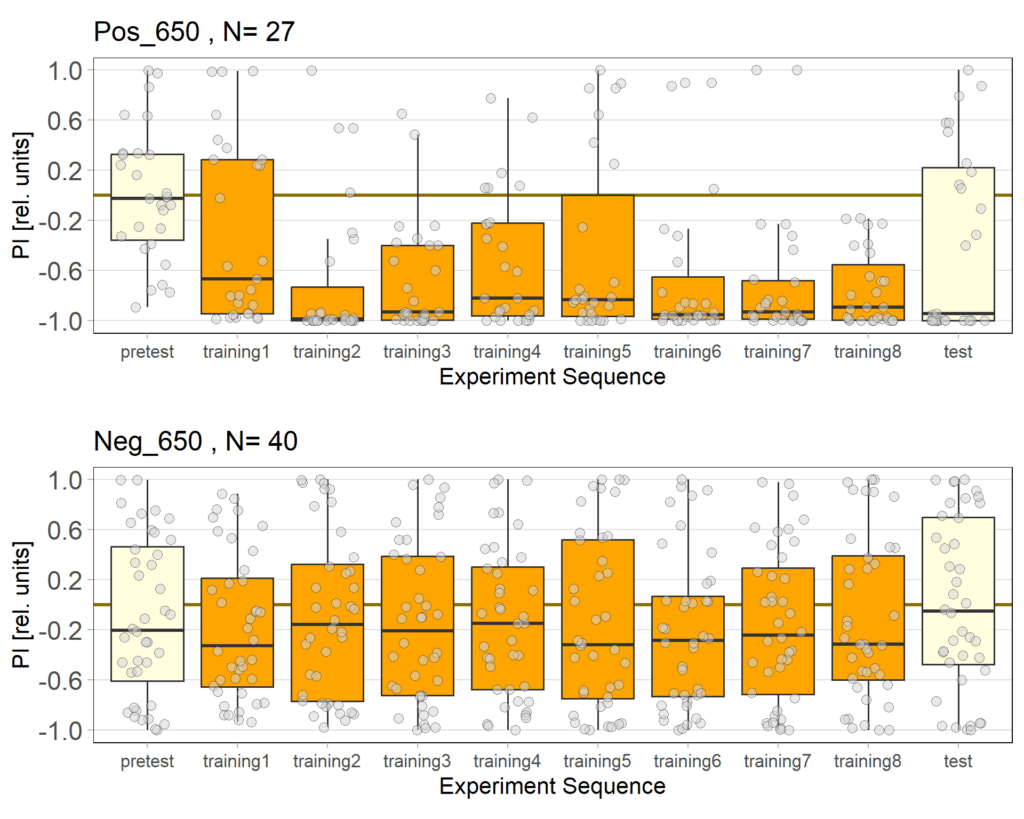
Alisa+Daniel experiments
Category: Optogenetics | No Comments
Yt learning_positioning effect
on Monday, June 30th, 2025 11:47 | by Julia Schulz
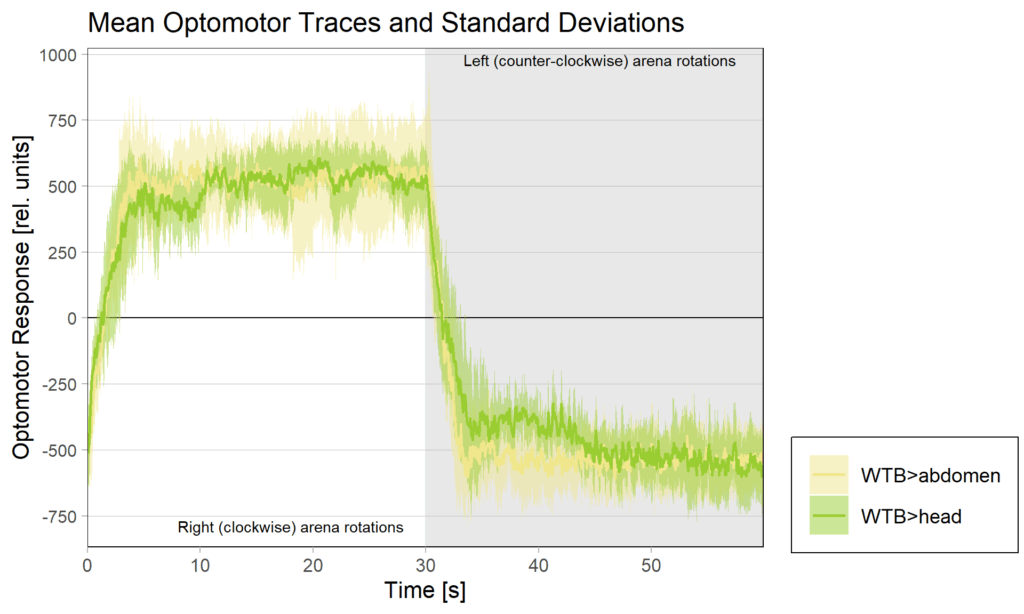
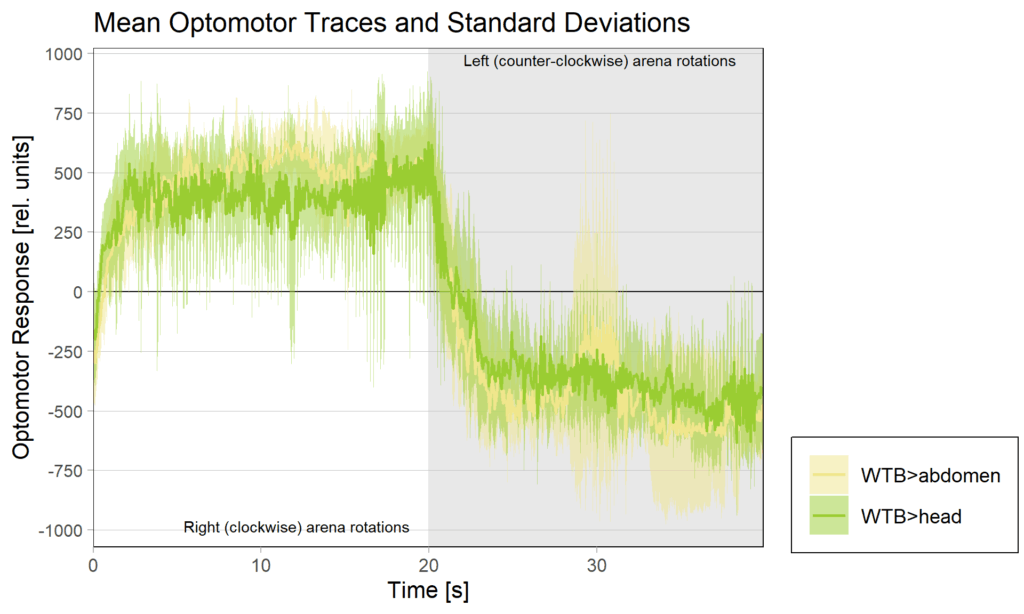
side bias can be compensated for by changing the positioning of the flies within the copper clamp from the medial to the lateral side facing away from the wall.
Category: Operant learning, operant self-learning, Uncategorized | No Comments
Final 3-Iodo-L-Tyrosine trial before testing PPM2 flies.
on Monday, June 30th, 2025 11:34 | by Daniel Döringer
I confirmed the action of the dopamine synthesis inhibitor 3-Iodo-L-tyrosine (3IY) in a previous trial, where treated flies showed decreased walking speed and distance when tested in an open field. In this trial I tested the flies “fresh out of the tube” so that they were able to feed on 3IY right to the point where they were tested. For the main experiment however, flies will be transferred from the food 2-5 hours before the experiment. To confirm, that the effect of 3IY treatment will persist also after this time span I moved flies from 3IY supplemented vials to fresh, empty vials at least 4 hours before measuring walking behavior in the open field. The results confirm, that the effect of 3IY is still present after this time.
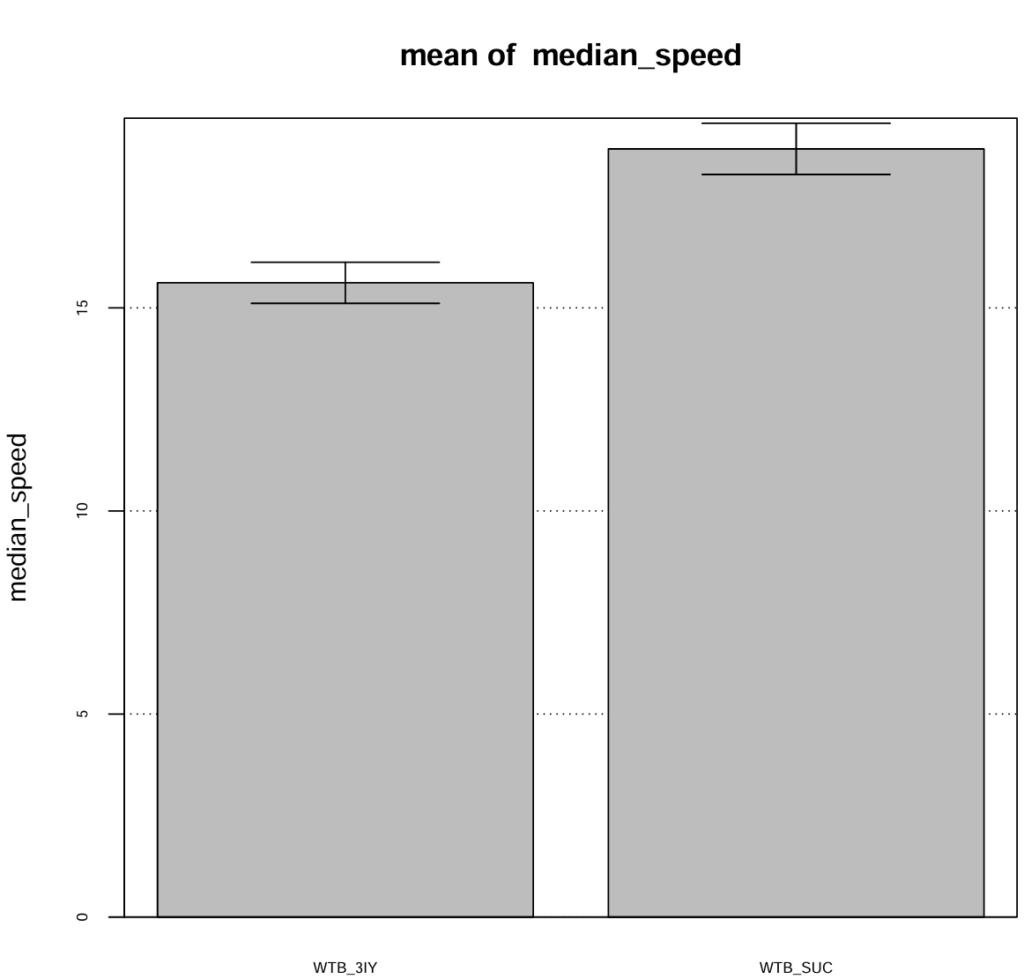
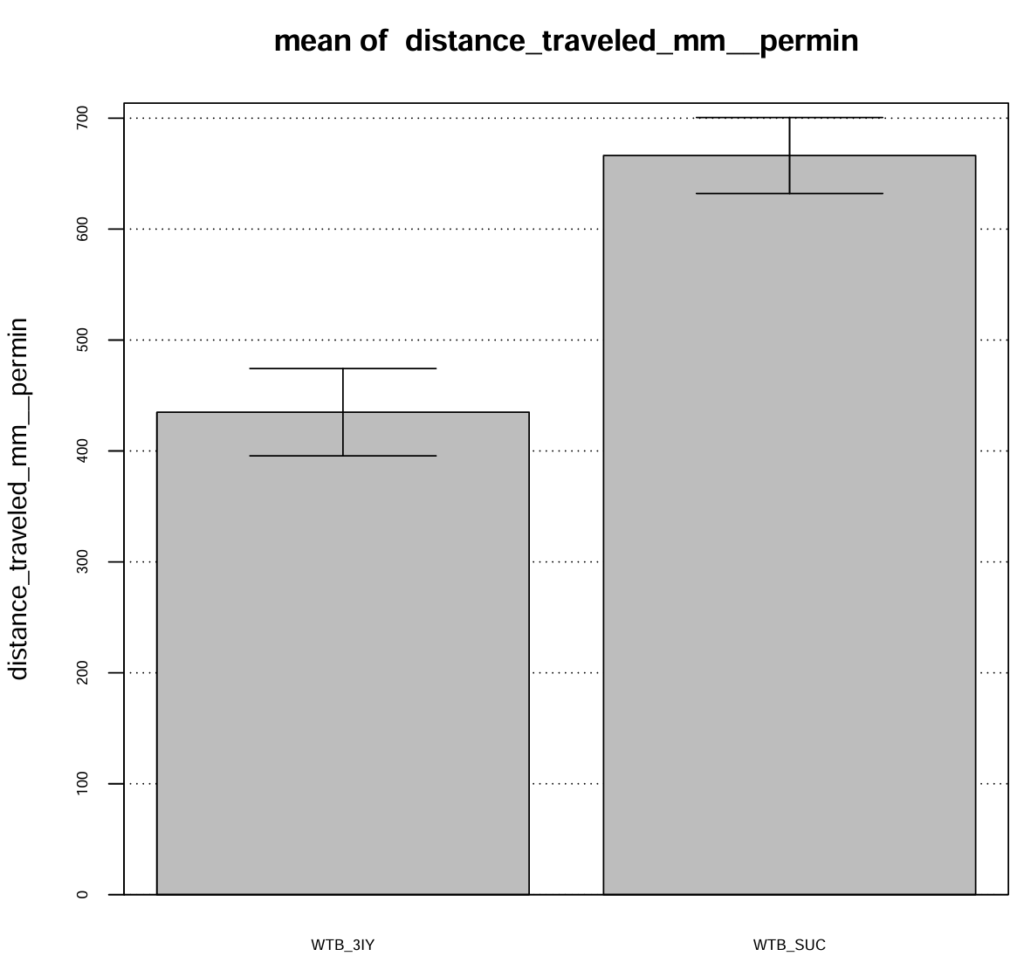
Category: Optogenetics | No Comments
Salt avoidance under blue and redlight
on Sunday, June 29th, 2025 10:10 | by Eva Schächtl
N=12
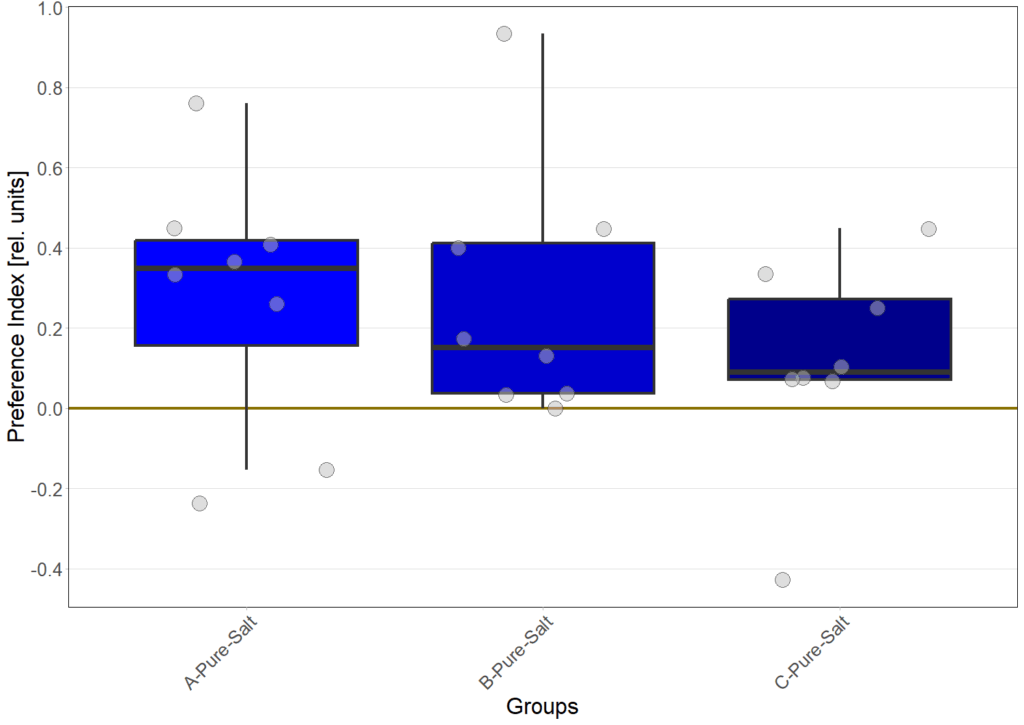
N=8
Category: genetics, Larve, neuronal activation, Optogenetics | No Comments
Temporal analysis of salt avoidance behavior in CantonS reveals deviations from established literature
on Tuesday, June 24th, 2025 5:16 | by Nils Henseling
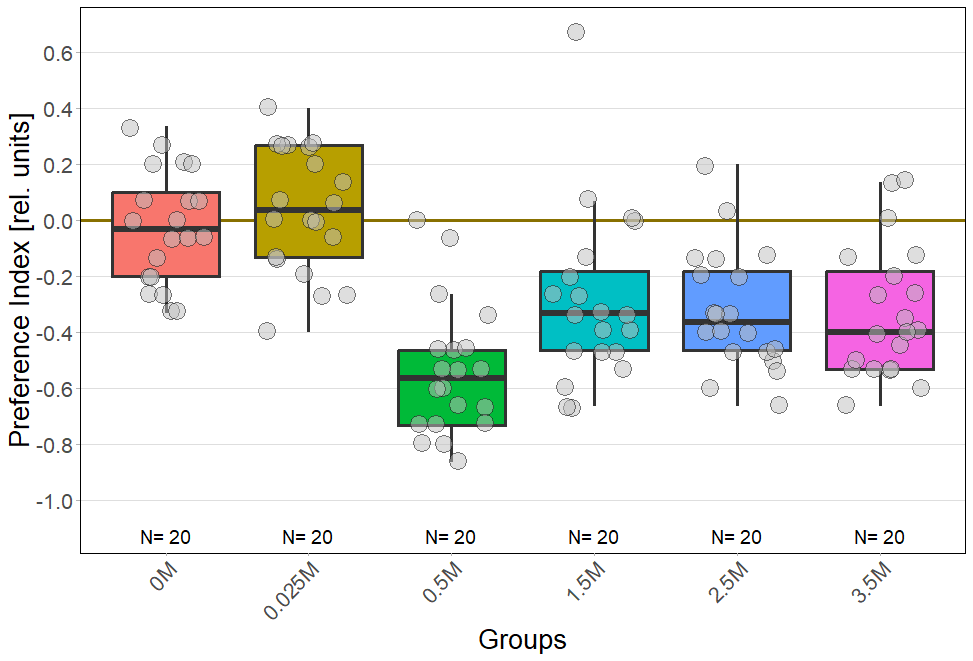
For every concentration: N = 20
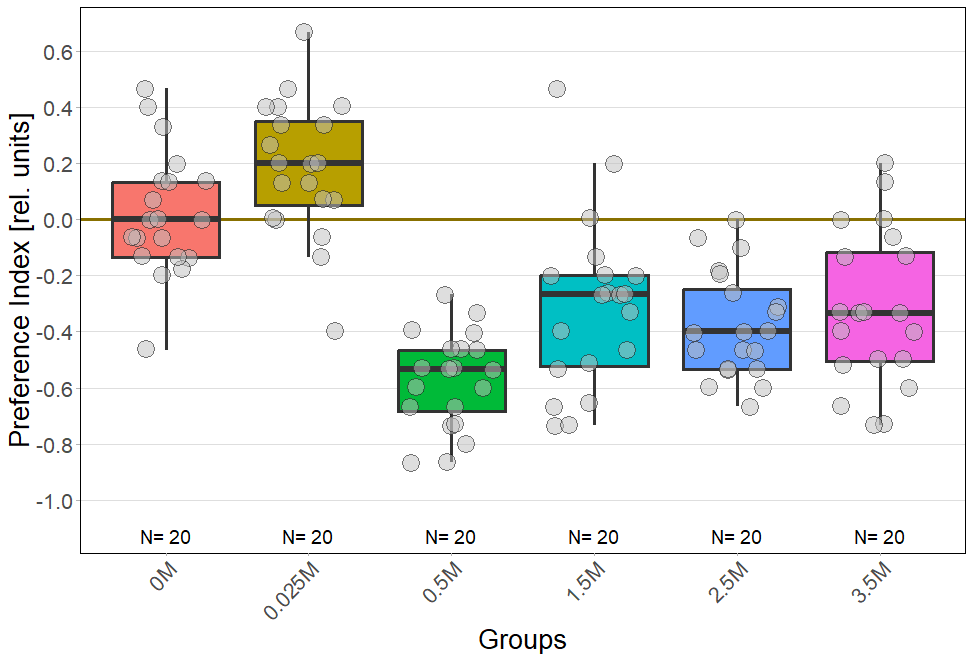
For every concentration: N = 20
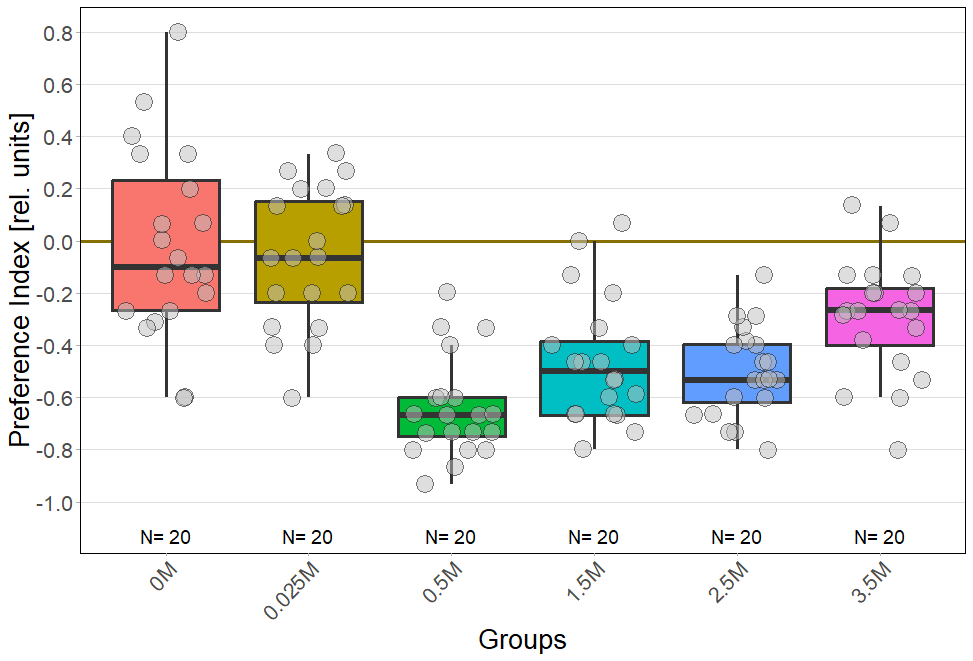
For every concentration: N = 20
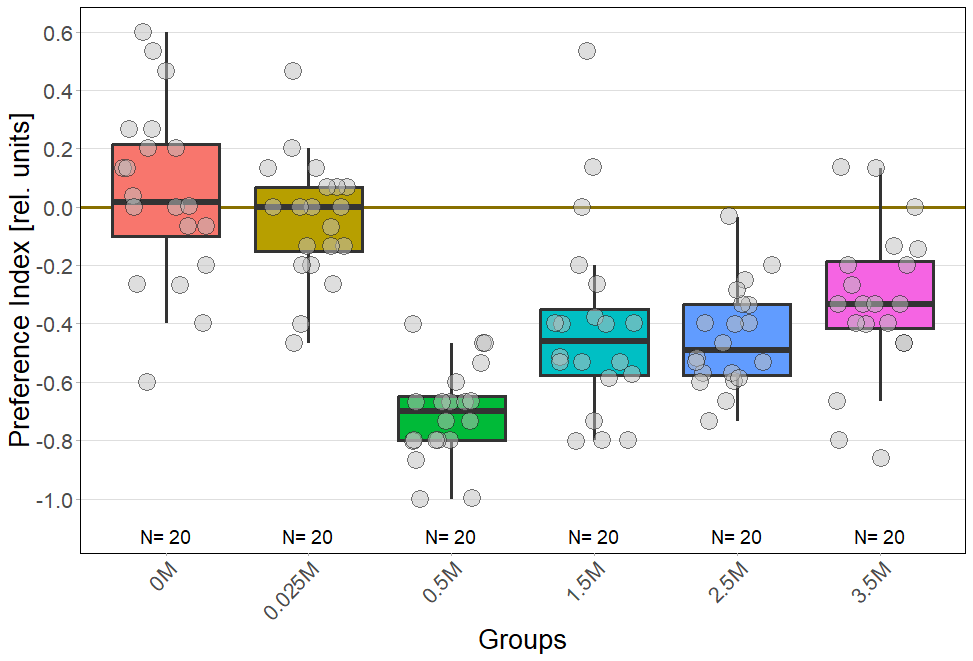
For every concentration: N = 20
Category: Food preference, Larve, Uncategorized | No Comments
YT learning_trouble shooting_body orientation
on Monday, June 23rd, 2025 1:39 | by Julia Schulz
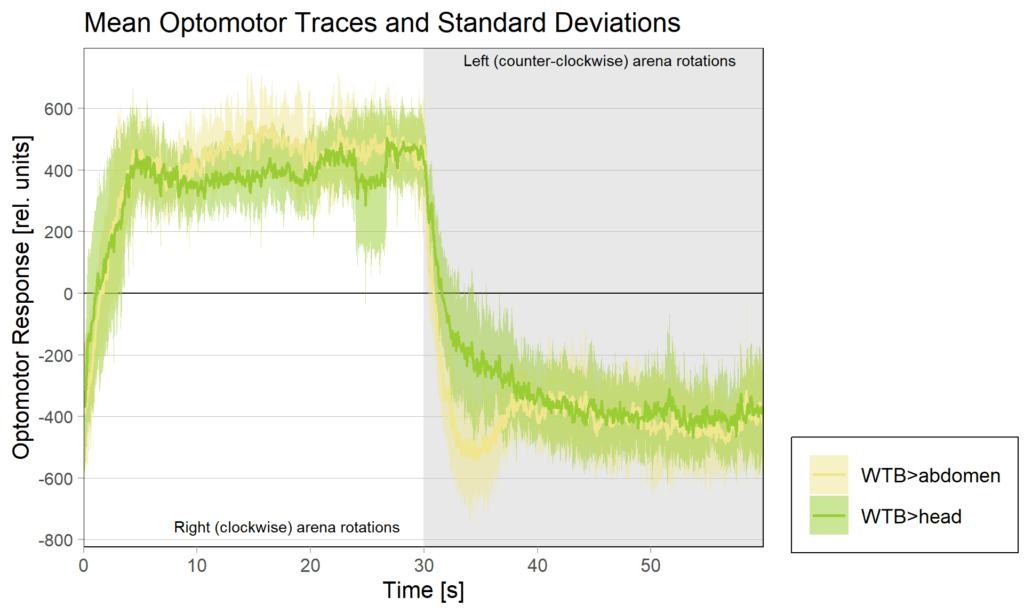
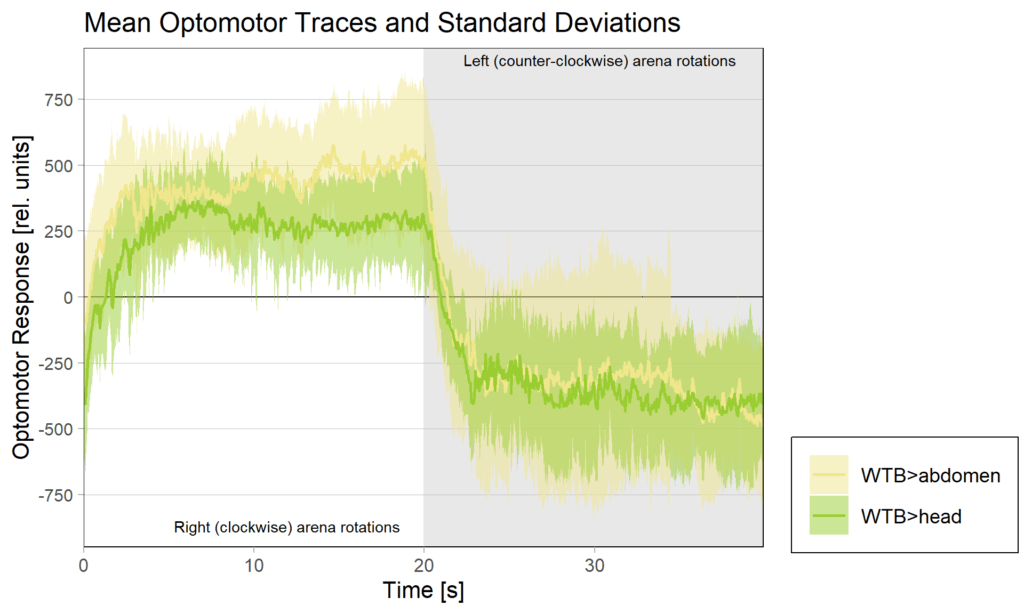
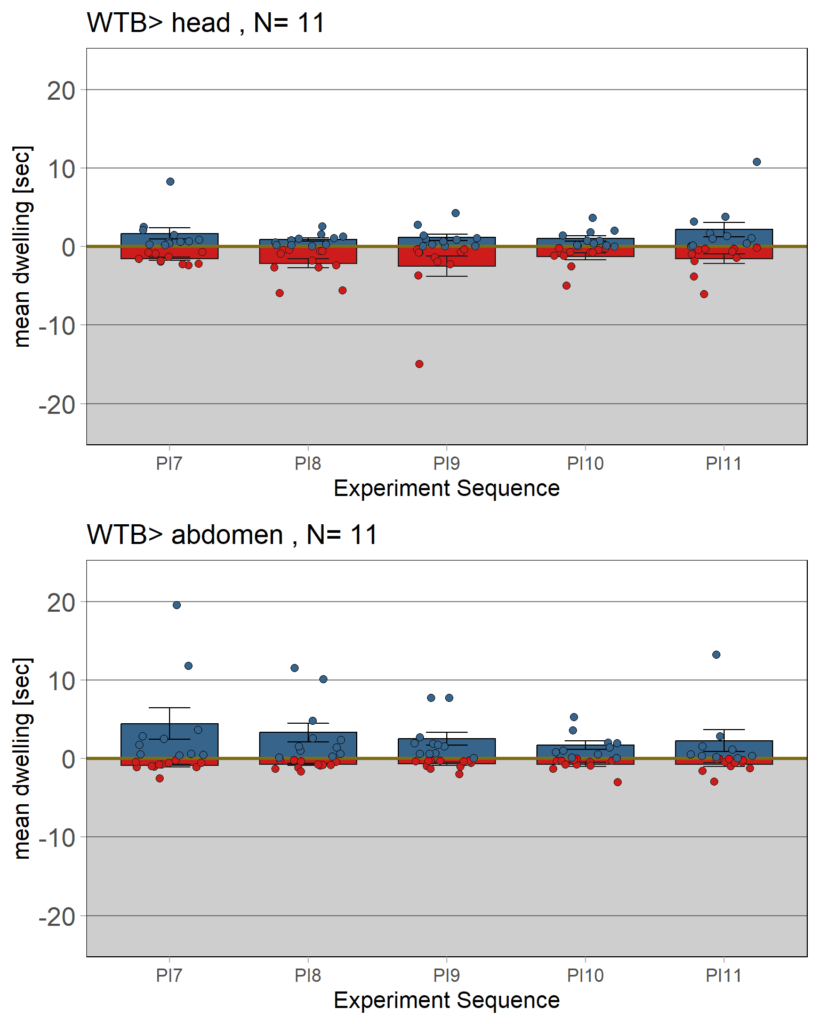
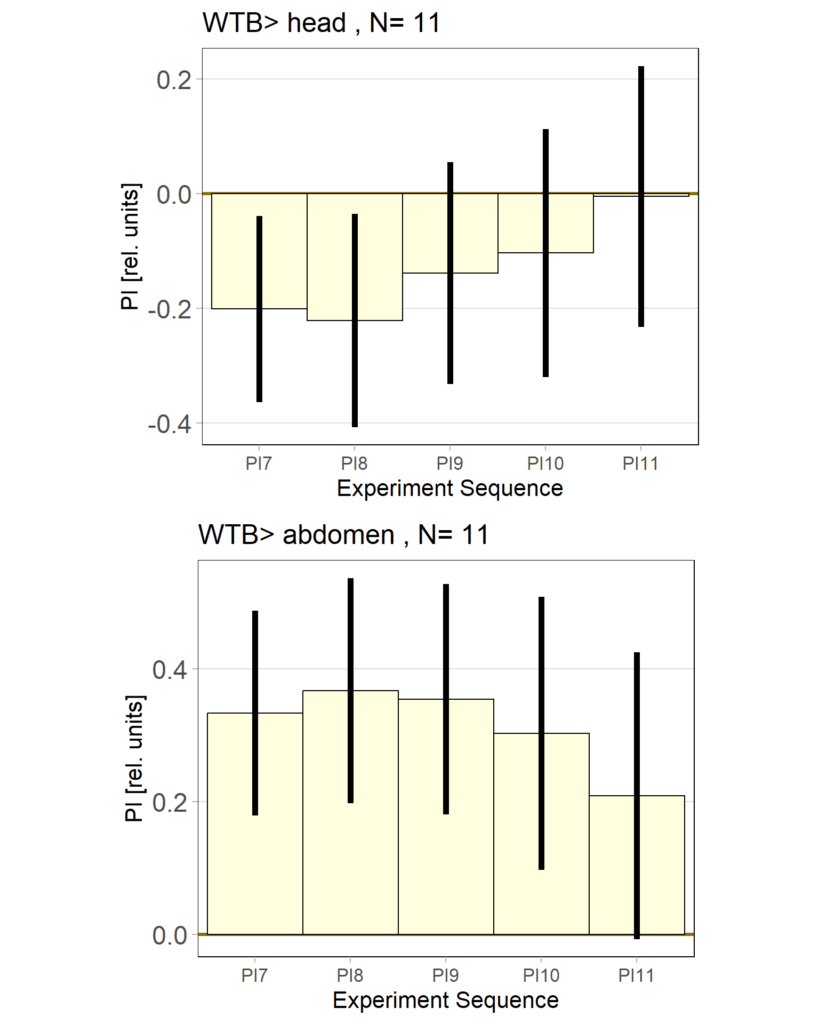
Category: Operant learning, operant self-learning | No Comments
Yt learning_trouble shooting
on Monday, June 23rd, 2025 1:37 | by Julia Schulz
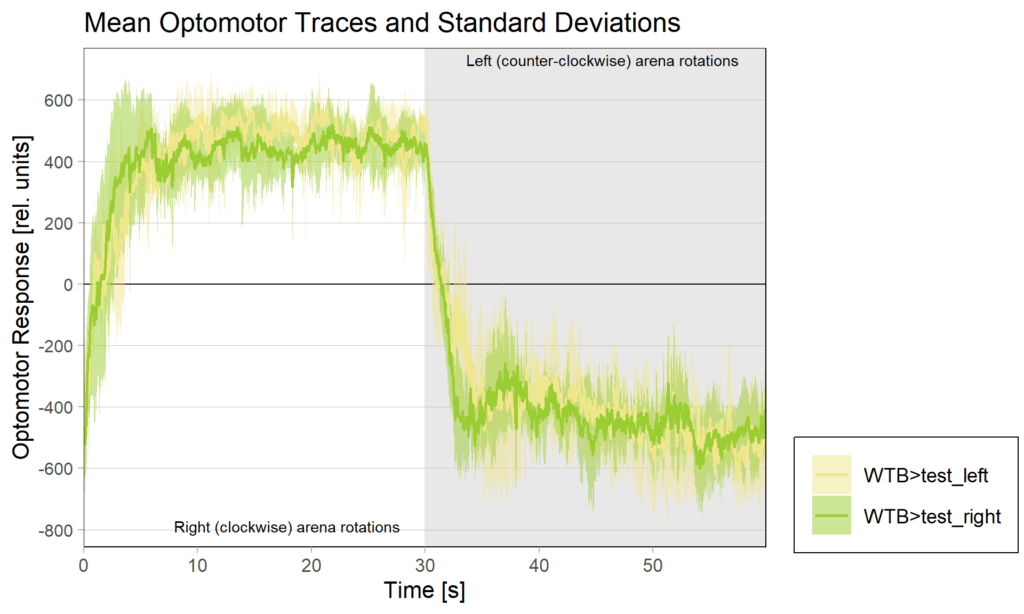
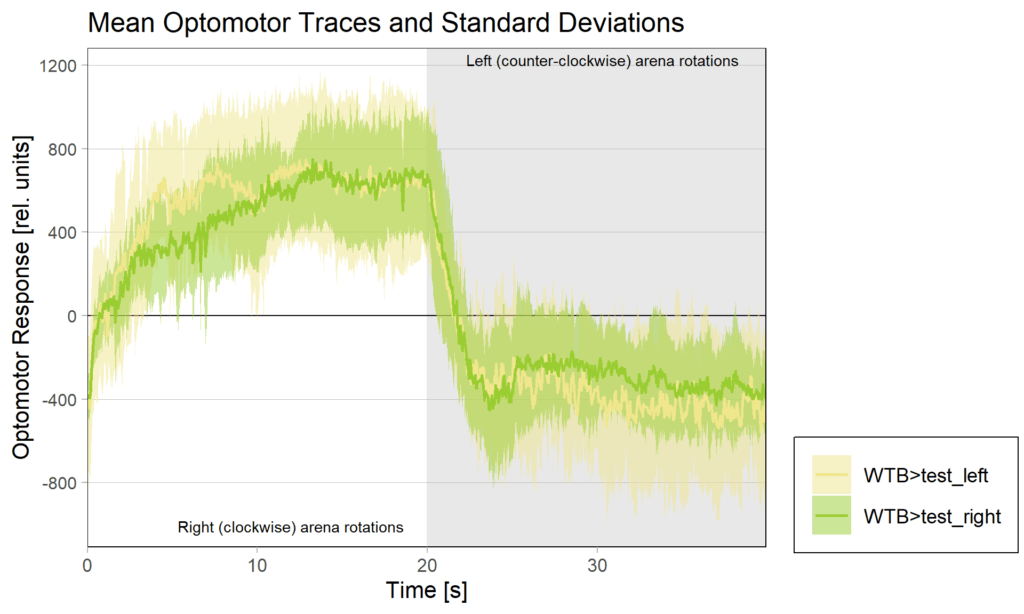
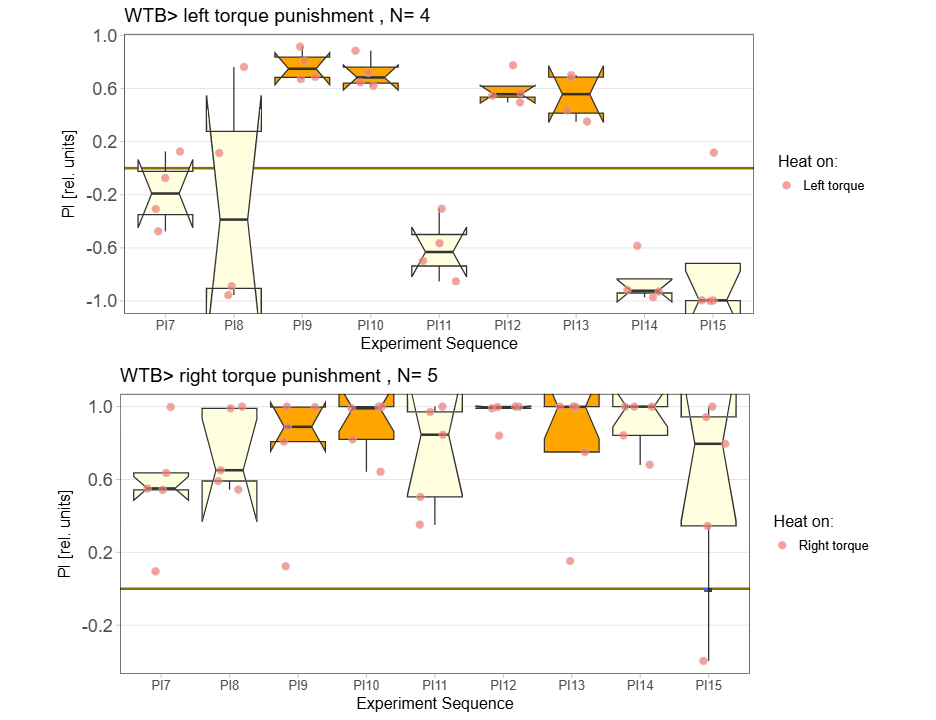
Category: Operant learning, operant self-learning | No Comments
3-Iodo-L-tyrosine Trial 3
on Monday, June 23rd, 2025 1:13 | by Daniel Döringer
For this trial I prepared 2mL of 3IY (10mg/ml in 5% Sucrose) 30 minutes before applying it to 8 pieces of filter paper stacked in the glass vial. I pipetted the whole volume of 2mL into the vial and discarded the excess liquid.
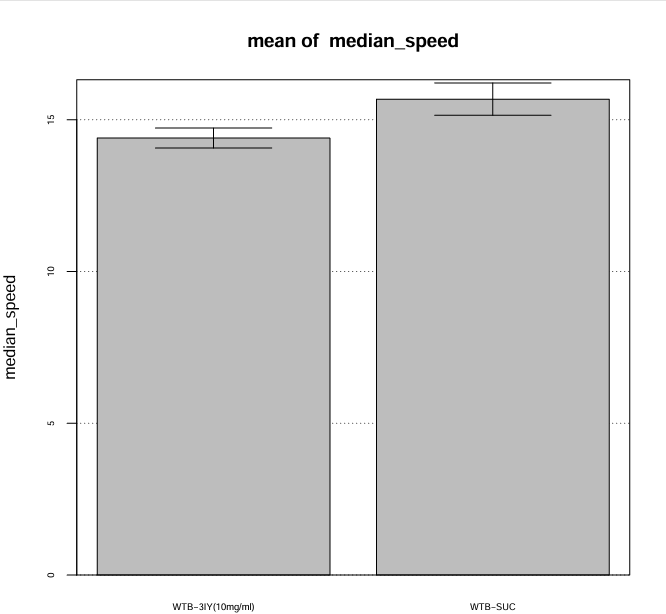
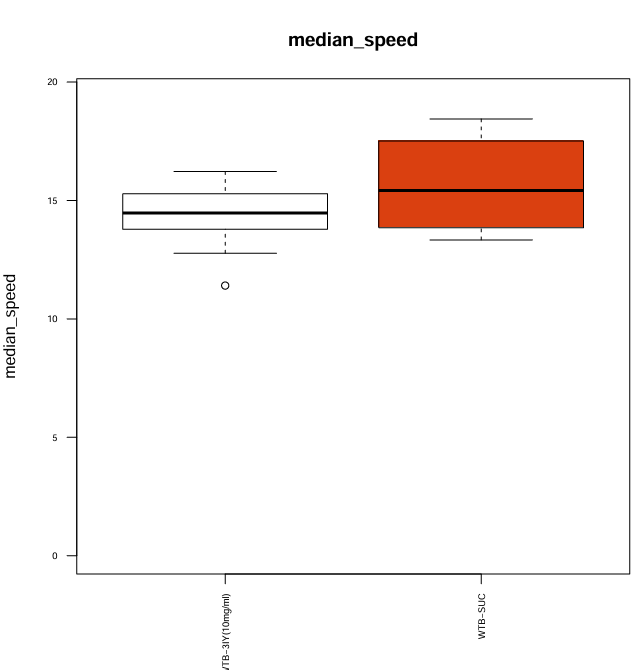
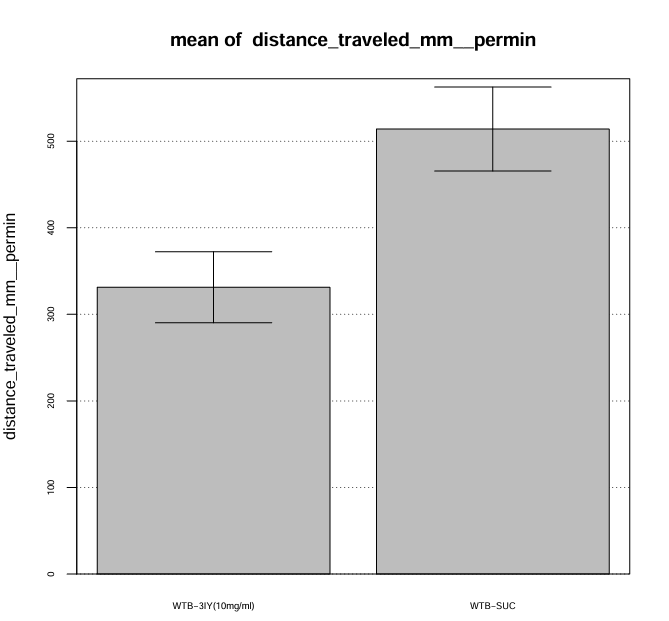
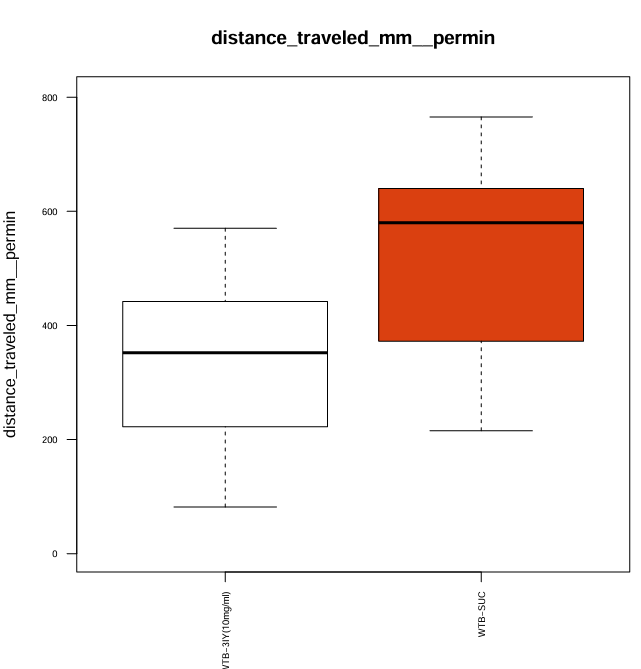
Category: Optogenetics | No Comments
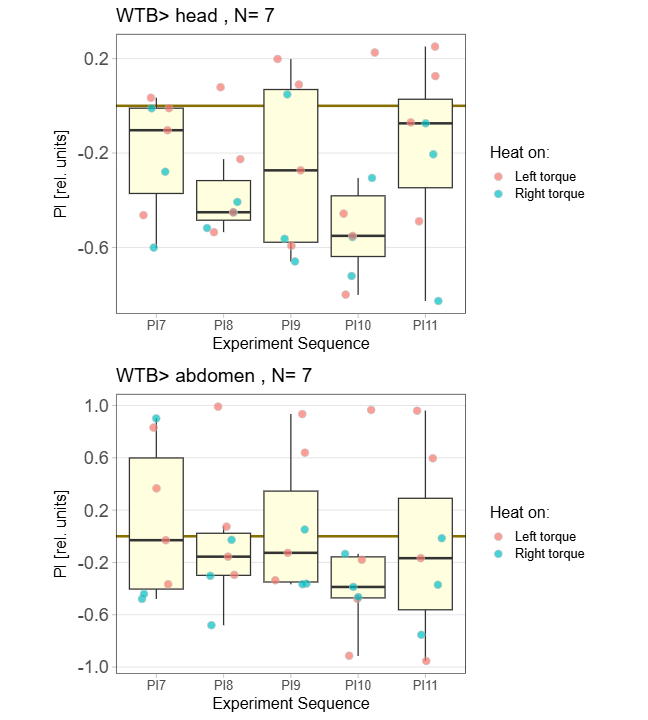
Trying to fix the Negativ Control for JoyStick Experiments
on Monday, June 23rd, 2025 1:06 | by Daniel Döringer
Recent results revealed problems with our negative control (Gr28bd-Gal4; TrpA1-Gal4+ EtOH) treated with EtOH. As the food for these flies was not substituted with ATR but only with Ethanol (ATR is dissolved in EtOH), the Chrimson channel should be non-functional in this group and the flies should therefore not avoid light. However, we observed negative PIs for this group. Since some activation of the Chrimson channel could be possible even without ATR, I tried to lower the used light intensity to values where the positve control would still avoid light, but the intensity would be too low for the Chrimson channel to activate when no ATR is present.
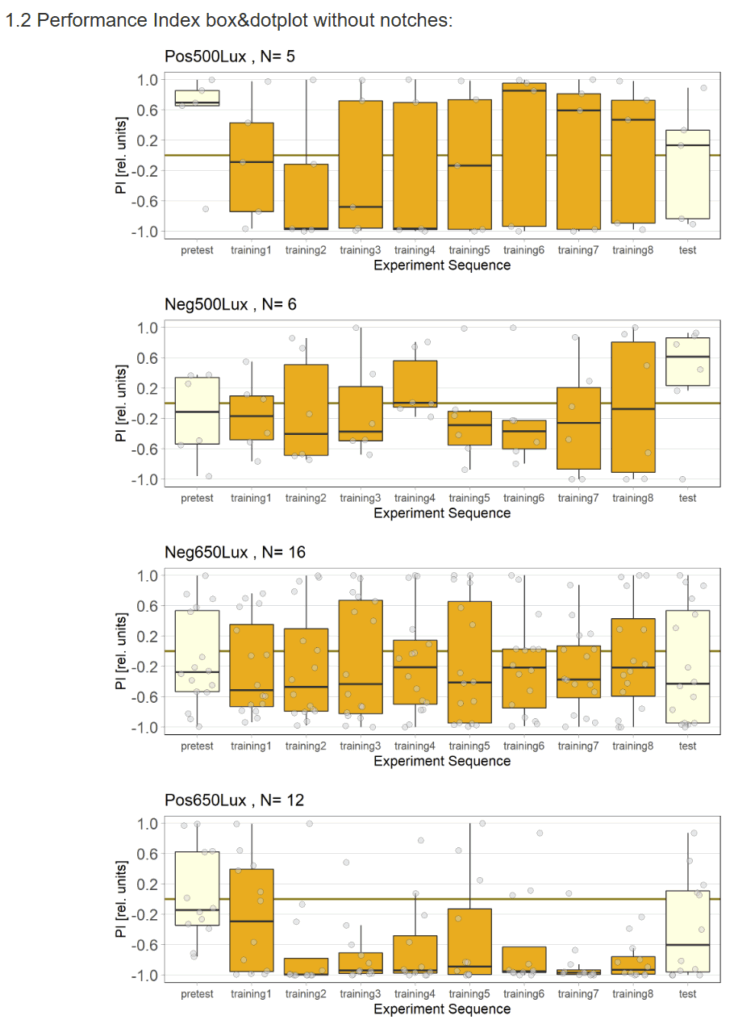
It seems that 650 Lux might still be too high. Therefore I will conduct another trial with 500 Lux.
Category: Optogenetics | No Comments
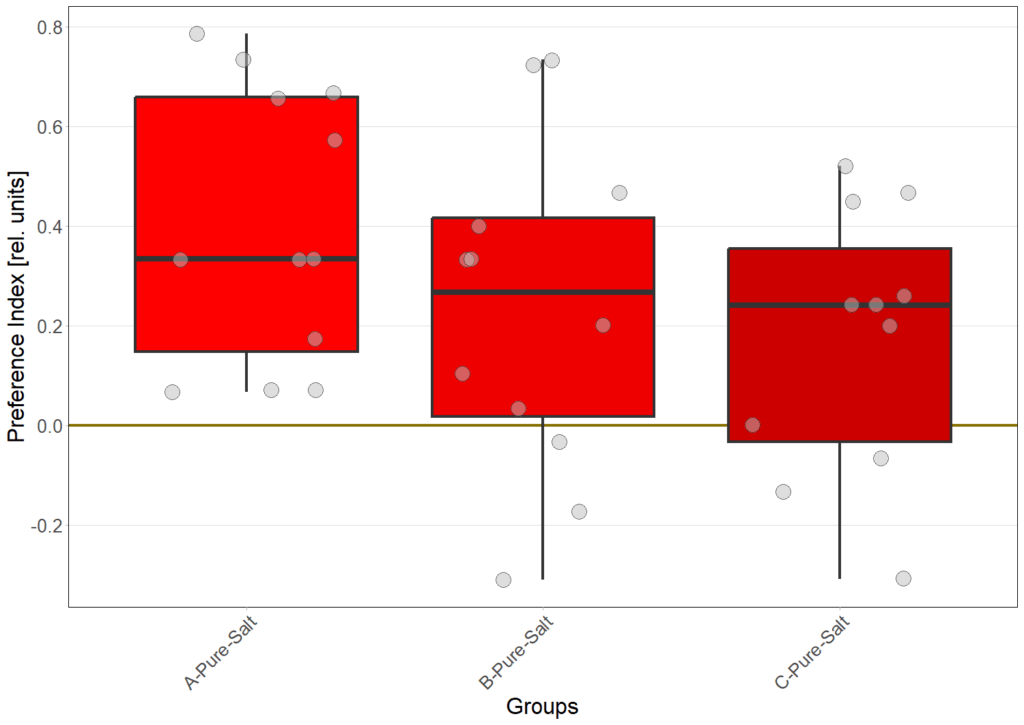
Gustatory preference final results
on Sunday, June 22nd, 2025 10:55 | by Eva Schächtl
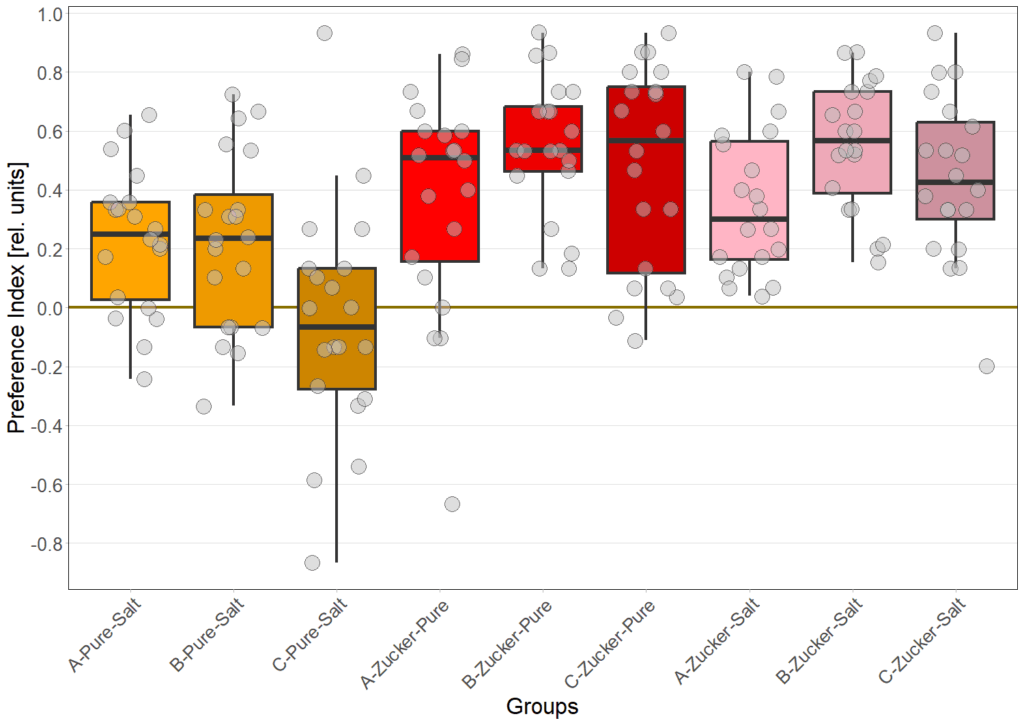
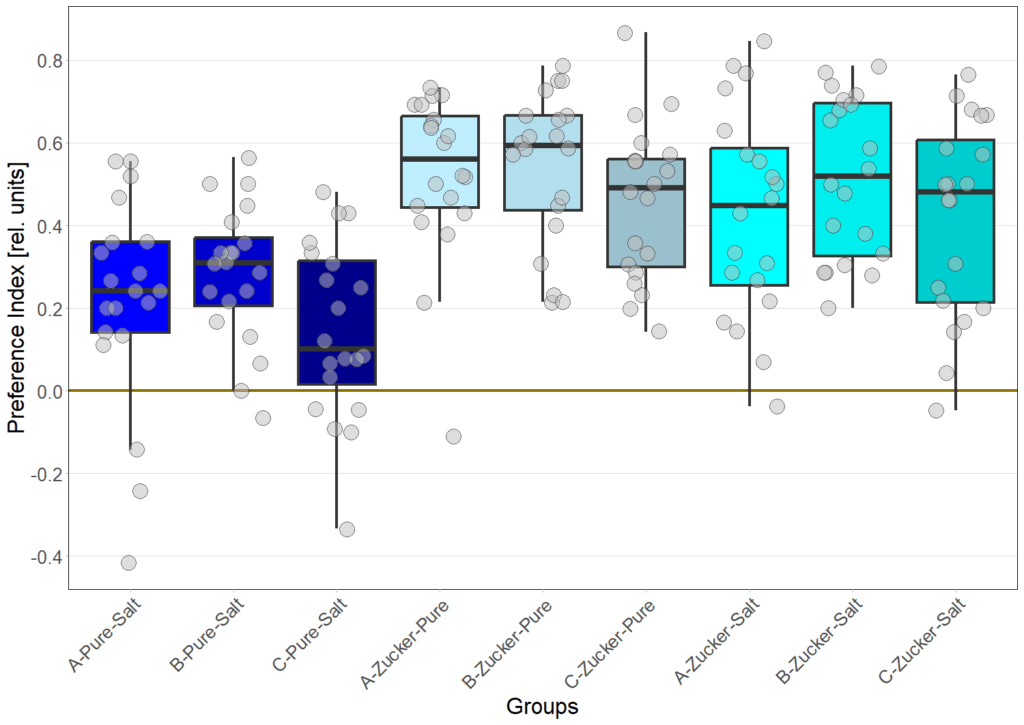
Category: Food preference, Larve, neuronal activation, Optogenetics | No Comments
