Better analysis of slope dependence on flight length and further insights in the SMAP code
on Monday, December 4th, 2017 1:02 | by Christian Rohrsen
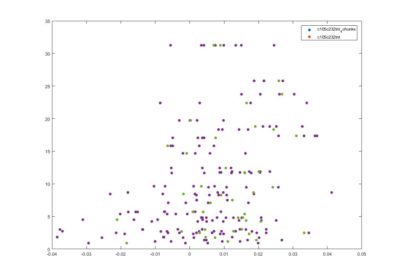
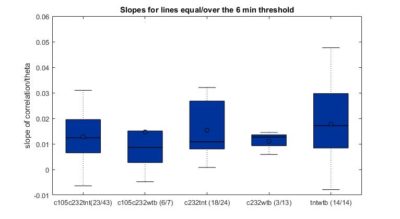
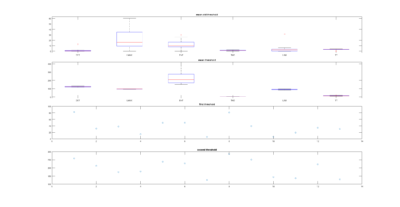
This is another way of showing how the slope of the SMAP analysis varies with length. I have choped the time series in 4 chunks and saw what was the slope for the chunks and for the whole time series. I think this is the best statistical way of doing it, it should not depend on what line was tested. I cannot see any effect.
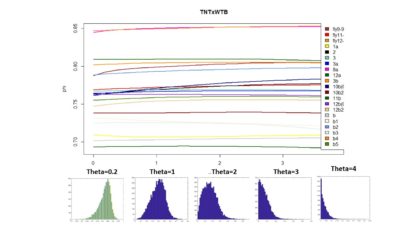
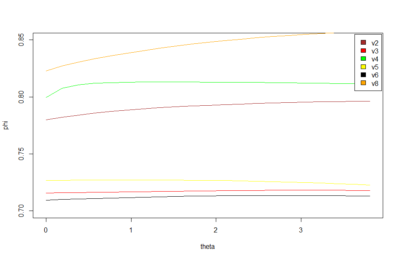
Here below is just to show what I found out in the code. What I thought that theta was controlling for nonlinearity in the model, for me it seems rather a control for under- overfitting in the model. So I might need to read the paper and see what do they say, and if it is the same as what I found out in the code.
Category: flight, Spontaneous Behavior, strokelitude, WingStroke | No Comments
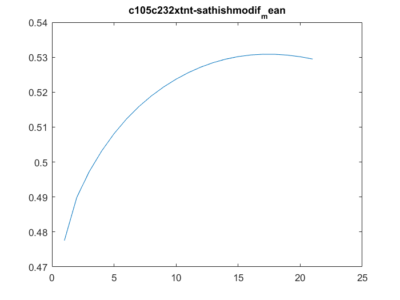
Do not agree with Sathish?
on Monday, November 27th, 2017 2:43 | by Christian Rohrsen
So this is the final graph assuming that 20Hz is the sampling rate. Sathish was not sure what it was and he said he will check.
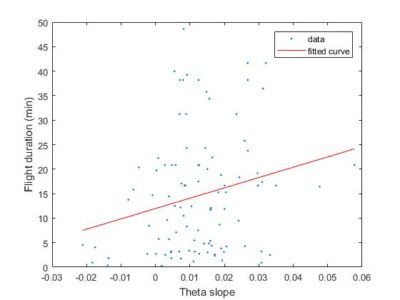
To have a better overview how the length of the flight compares with the slope obtained from the SMAP. There is no big correlation whatsoever with around 100 flies. Sathish found this in his thesis with data from seventy something WTB, but to me it seems like an anecdotal result.
Here the same as above, just for showing the fit line.
Category: flight, Spontaneous Behavior, strokelitude, WingStroke | No Comments
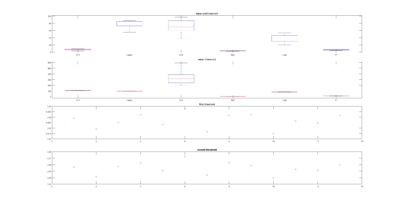
All sathish data analysis
on Monday, November 20th, 2017 1:47 | by Christian Rohrsen
This are the results of all of the flies analyzed from Sathish. I just need to know the frequency of the acquisition to exclude the ones that are below 6 minutes.
Category: flight, Spontaneous Behavior, strokelitude, WingStroke | No Comments
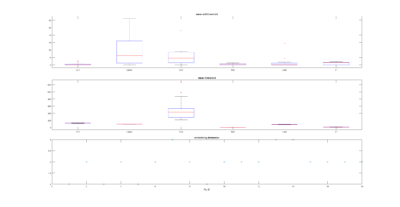
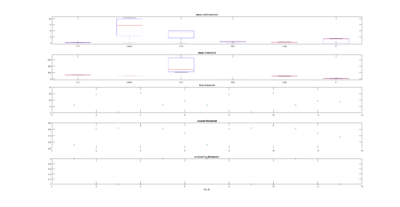
Sathish data
on Monday, November 6th, 2017 1:37 | by Christian Rohrsen
I have analyzed the c105+c232 > tnt data from Sathish. There are a total of 43 flies, although many of them only flew for a few minutes, and therefore should be discarded. Below all of the individual fly scores. I need to analyse now the other groups. This is btw the modified data set, whatever that means for Sathish.
This is a video of the projection from the torque data from Maye et al. 2007. The spikes are not that well sorted in this case as in the Strokelitude. I guess this is because the spikes do not look so smooth.
Category: flight, Spontaneous Behavior, strokelitude, WingStroke | No Comments
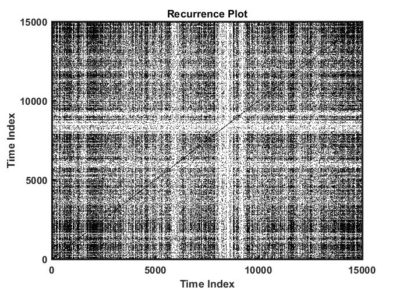
Recurrence quantitative analysis
on Monday, October 30th, 2017 11:43 | by Christian Rohrsen
This is an example of a recurrence plot analysis. In the first graph is shown in single point in time in the optimal embedding dimension and the distance to the other points. For the recurrence plot analysis it is needed to put a threshold to make it binary. This is the second graph. From this second graph one can count many parameters like determinism, laminarity and so on. From what I see, the plots from the Strokelitude as well as Bjoern´s flight simulator in Maye et al 2007 show similar pattern (kind of crosses with vertical and horizontal lines).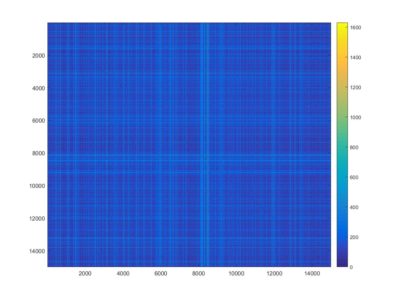
This is a measure of the Recurrence Quantitative Analysis of different groups. Recurrence threshold is a tricky and to some extent subjective measure, so this is why I tried two different ones.
DET: recurrence points that form a diagonal line of minimal length, the more diagonal, the more deterministic.
LMAX: Max diagonal line length or divergence. Sometimes considered as an estimator of max. Lyapunov exponent
ENT: Shannon entropy reflects the complexity of the system
TND: info about stationarity (trend)
LAM: Laminarity is related to laminar phases in the system (intermittency). It is tallied as vertical lines over a threshold.
TT: Trapping time, measuring the average length of vertical lines. Related to laminarity.
Automat
Openloop
Uniform
Category: flight, Spontaneous Behavior, strokelitude, Uncategorized, WingStroke | No Comments
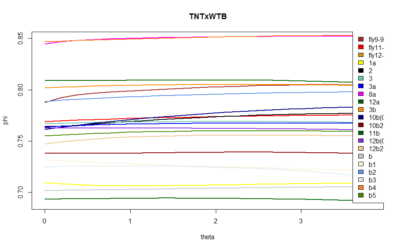
SMAP results
on Monday, October 9th, 2017 2:34 | by Christian Rohrsen
These are the results of the SMAP for the TNTxWTB. I also have done a few for the c105;;c232xWTB but there is not much to say. I would say that the cleanest lines show a bigger slope, but prone to subjectiveness.
In addition, I have done some animations of the attractors that I have posted on slack because of size.
Category: flight, genetics, R code, Spontaneous Behavior, strokelitude, WingStroke | No Comments
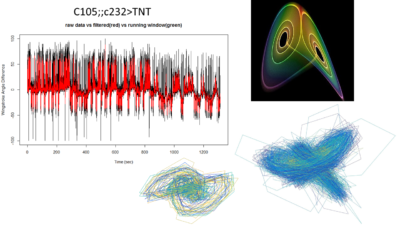
Attractors for c105;;c232>TNT
on Monday, October 2nd, 2017 5:35 | by Christian Rohrsen
This is what I showed about one fly from this line showing the attractor.
This graph is what I forgot to show in the lab meeting. There are the 6 best traces from this same line. All of them selected ad hoc subjectively. The three best of them to my eyes are exactly the three above in the graph (v8-this one is the one shown in the picture with the attractors-,v4,v2). What does this means? the ones with better traces (subjectively) have higher offset in the phi, this means that they are more predictable overall (maybe because more resolution?). In addition, they show higher slopes which means more nonlinearity.
Category: flight, Spontaneous Behavior, strokelitude, WingStroke | No Comments
Measuring Wingstroke Amplitude with Strokelitude (V)
on Monday, December 14th, 2015 2:42 | by Pablo Martinez
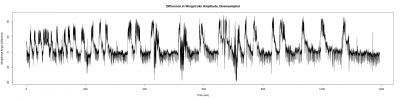
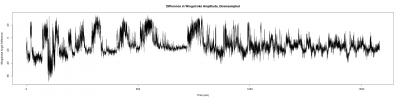
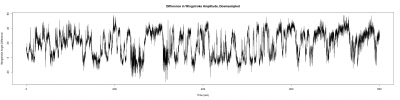
For a further data analysis, we should have a minimum number of samples. During the last week, I have been measuring the wingstroke amplitude of the flies, to get at least 10 samples of each one(two controls and the experimental line). Here three examples of the different lines:
Males WTB x C105;;C232 (control)
Males UAS-TNT-E x C105;;C232
Males UAS-TNT-E x WTB (control)
Category: flight, Spontaneous Behavior, strokelitude, WingStroke | No Comments
Measuring Wingstroke Amplitude with Strokelitude (IV)
on Monday, December 7th, 2015 2:56 | by Pablo Martinez
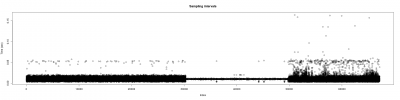
After getting good results in the measurement of the wingstroke, and solving problems with the sampling intervals (image below). I have started to measure flies for the experiments.
The time between samples was different depending on the background programs running on the background:
Figure 1
More differences in time within samples when more programs are running at the same time as strokelitude (3nd 1/3 of the plot), when just the display of the camera is running( 1st 1/3 of the plot), and everything shut down(2nd 1/3 of the plot).
The differences among sampling intervals was bigger but with an adjustment of the data, Christian Rohrsen managed to changed. The time between two samples could arise until 1.7 seconds and with the correlation, the time is not bigger than 0.05 seconds.
The strains of flies used fore the experiments are:
UAS-TNT-E, that express the tetanus toxin in the neural cells, using GAL4 system. Being Used as a control.
c105;;c232, that contains a promotor region to express the toxin. Being used as a control.
And the cross between both to have the expression of the tetanus toxin.
Some data from the control flies (UAS-TNT-E):
And some data of the spikes:
Category: flight, Spontaneous Behavior, strokelitude, WingStroke | No Comments
Interpolations and Spike analysis
on Monday, December 7th, 2015 2:42 | by Christian Rohrsen
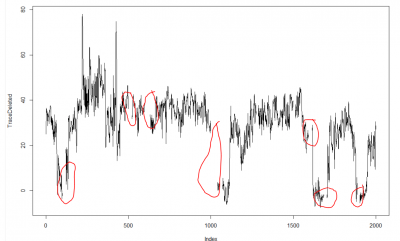
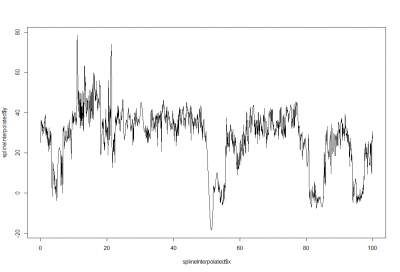 This is just a proof of concept how useful the interpolation is for this purpose: and I would say it´s almost useless. If I delete from the raw data (1st graph) some pieces (2nd graph), and I make a spline interpolation (3rd graph) it doesn´t match that well. Linear interpolation was done in previous weeks with not much of a success. Anyway, the result of a linear interpolation can be imagined by eye just by joinning the two ends with a straight line, and this doesn´t occur in the fly behavior as we can see in the raw data (1st graph).
This is just a proof of concept how useful the interpolation is for this purpose: and I would say it´s almost useless. If I delete from the raw data (1st graph) some pieces (2nd graph), and I make a spline interpolation (3rd graph) it doesn´t match that well. Linear interpolation was done in previous weeks with not much of a success. Anyway, the result of a linear interpolation can be imagined by eye just by joinning the two ends with a straight line, and this doesn´t occur in the fly behavior as we can see in the raw data (1st graph).
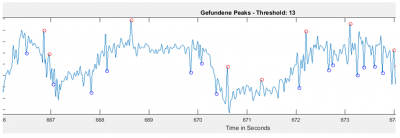
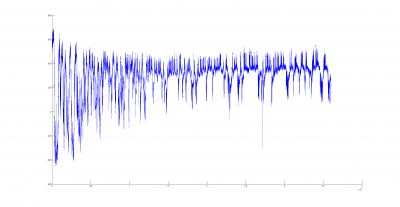
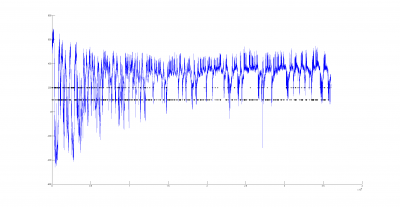
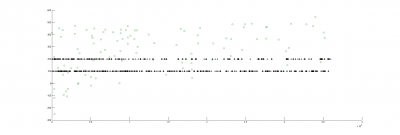 There could be two posibilities for spike detection: one is the one from Ute and the other one is the one from Maye. It seems to me that the one from Maye is more precise. I have run the script but I do not get so many spike detections as he gets. I did try several thresholds for spike detection and doesn´t change very much. So I have to work more on it to see what is really the important factor for a proper spike detection
There could be two posibilities for spike detection: one is the one from Ute and the other one is the one from Maye. It seems to me that the one from Maye is more precise. I have run the script but I do not get so many spike detections as he gets. I did try several thresholds for spike detection and doesn´t change very much. So I have to work more on it to see what is really the important factor for a proper spike detection
Category: flight, R code, Spontaneous Behavior, strokelitude, WingStroke | No Comments