automatic uplaod of data. step2
Figure of result, data, raw data and code uploaded automatically. Still need to get a way to get the statistical analysis published along with it.
I will try to make a video to highlight how fast and easy it is once the code is written.
Bjoern, when do you want to present this? Do I have time or should I do it tomorrow night ? :-)
the R code was made public: everyone can access it without login
bloomington flies arived
3 stocks came dead(26167 Cyk, 8641 da-Gal4, 34716 PKC53e RNAi). The other are good, although one was weak.
(7018 tubGal80ts on 3d, with cyo on 2nd
4440 201y
28997 TNT 3
27491 PKC53e RNAi
28837, and 28838 tnt2)
Automatic upload of data on figshare: step1
https://figshare.com/articles/PKC53e_putative_mutant_in_self_learning/97792
This was 100% uploaded directly from R.
First step toward automatic upload of data accomplished!
more permanent link, I think: https://dx.doi.org/10.6084/m9.figshare.97792
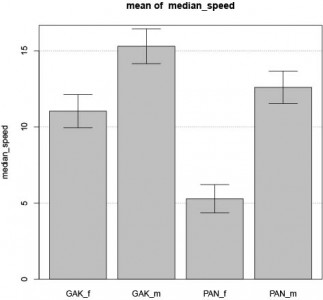
LiLiu data on buridan
I started to work with the data I got from Li Liu. I have no metadata file, so I am not sure what will finally come out of the analysis.
What is already clear is that the number of female and male they tested seem different for each group and there is a big difference in speed between male and female… The value reported are also huge (>20 mm/s, no distinction male/female) while I get the normal 15mm/s (if the data I got is in mm and not in pixel, of course).
I do not know what GAK and ‘PAN stands for…
Remember that from our data, there is no significant difference in male versus female speed.
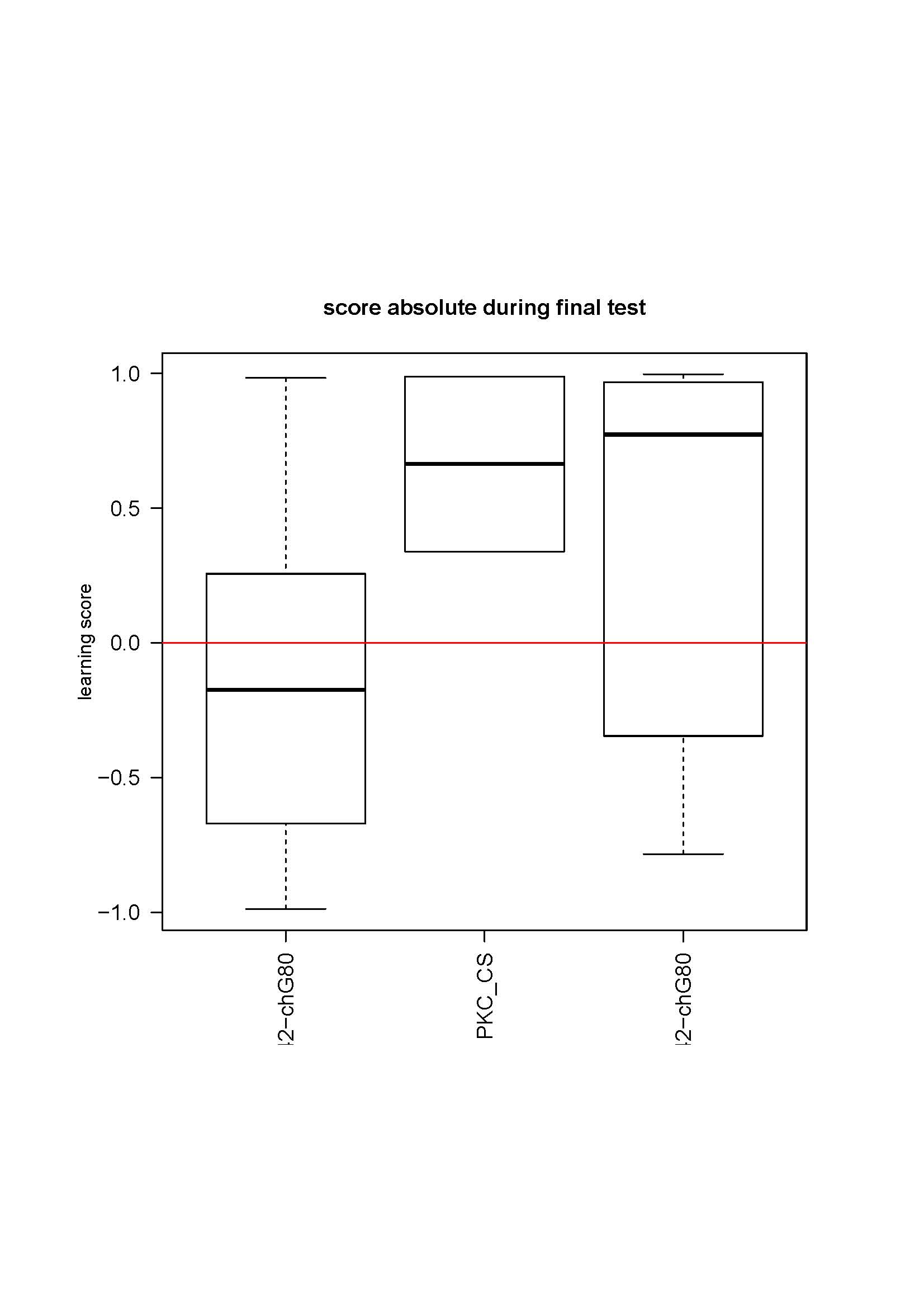
c232,105yx UAS-TNT self learning
none of the group learn. Flies are quite weak. The two groups with UAS show a clear problem in operant behavior (low score during learning phases)…
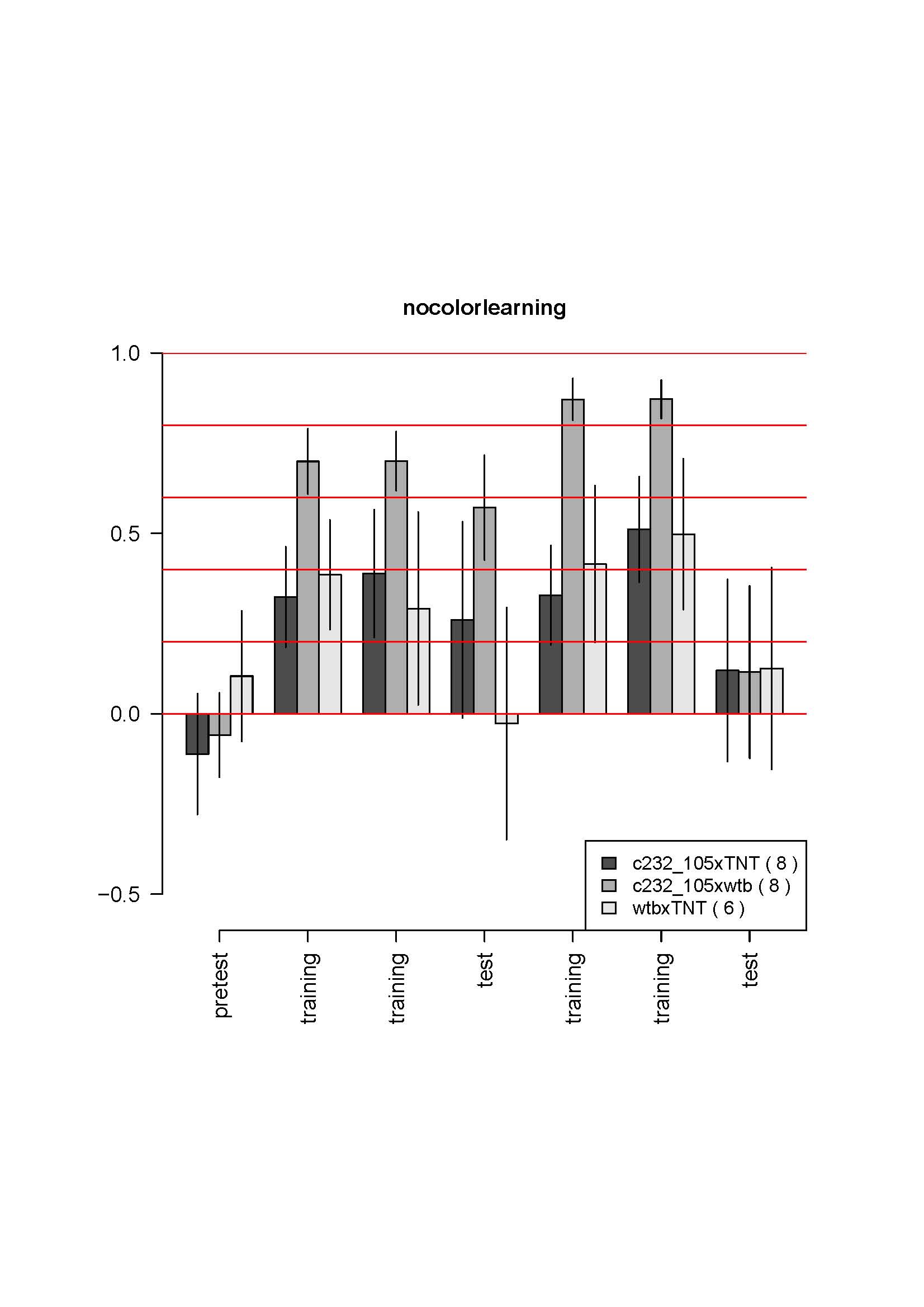
d42Gal4,chaGal80 (follow up)
Here is the data for first two crosses:
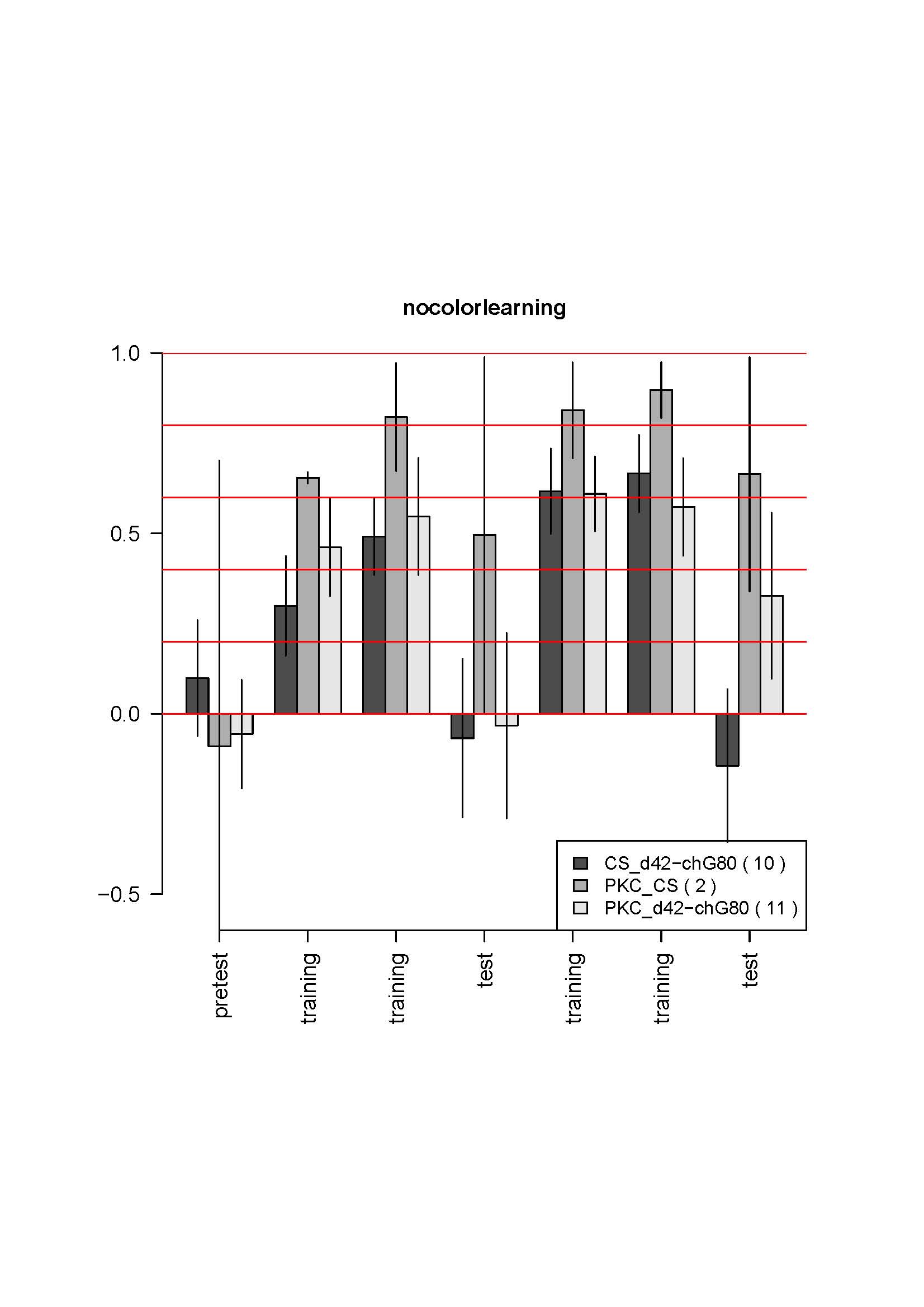
No changes from the first 4-6 tests. The most striking is the absence of learning for the control. It does not make sense. I also controlled that the genotypes were not inverted by looking at GFP fluorescence which is much weaker in the test line (due to the tubGal80ts presence) than in the control line. Could it be an effect of GFP presence in some neurons? The CS strain should be unguilty, since the CSx c380 had no phenotype in the previous cross…
A third cross is coming, with the elavGal4 control, maybe it is only bad luck… But the results were so reproductible so far, and so solid… it is strange. I got the parental lines out of the stock to get fresh flies for the next cross.
I should also cross new flies now. I am planning these groups (male-female):
UGFP;;d42gal4,chaGal80 x tubGal80ts,U-PKCi
CS x tubGal80ts,U-PKCi
UGFP;;d42gal4,chaGal80 x CS
UGFP,c380;;,chaGal80 x tubGal80ts,U-PKCi
Torque distribution
I had seen beautiful bell shaped distribution of torque (around 0) from the Heisenberg’s lab. We thus checked the data we have (the 6 minutes data we produced with the same flies for the torque meter and compensator. Data produced on the same day, or later (when sathish was mastering the preparation a bit better).
Here is the distributions:
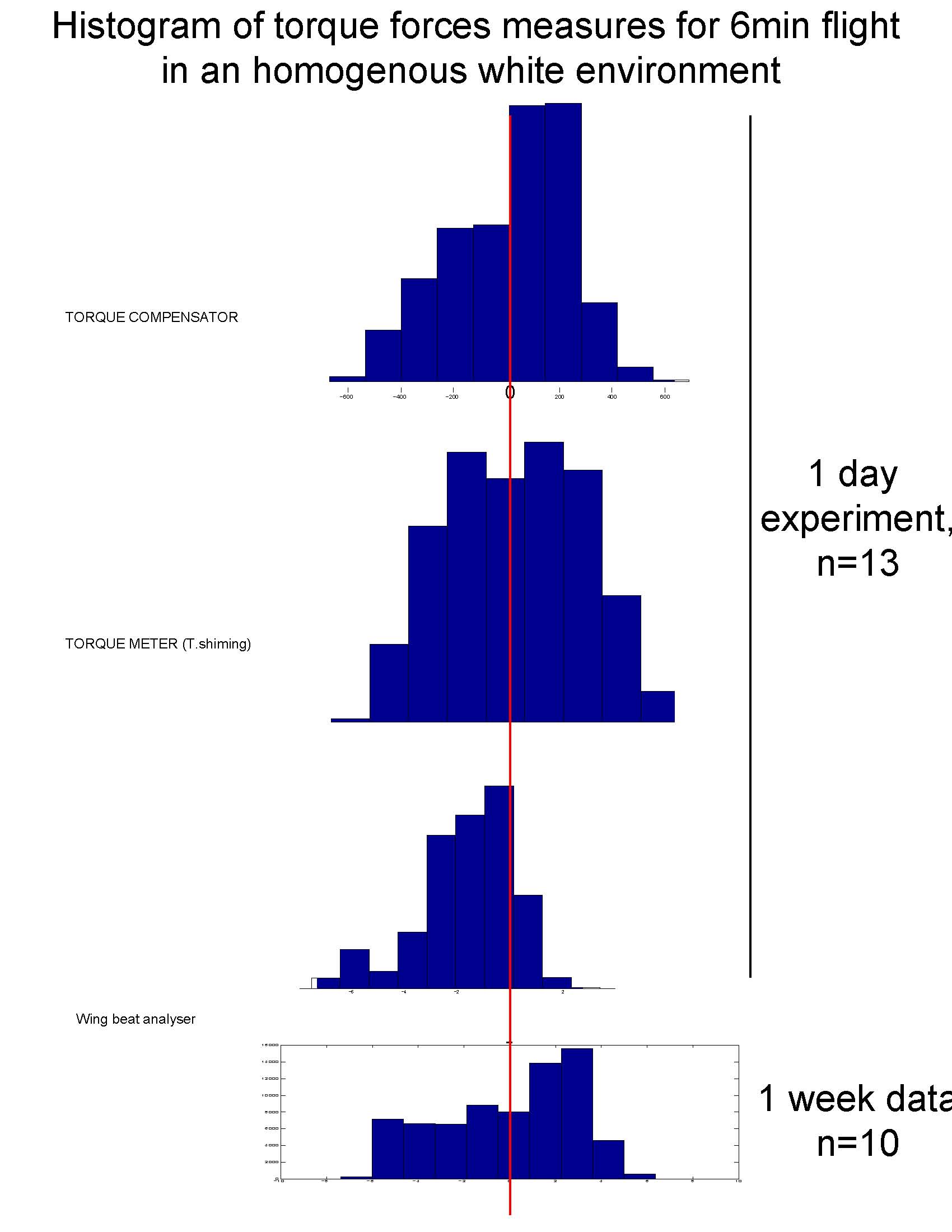
Our distribution are close enough to the bell shape obtain by the Heisenberg’s group. The wing beat analyser seem to lead to different torque calculation, though.
PS: no difference seen in the frequency of spikes on the other hand.
SMap before and after self-learning: no difference
small but significantly different from 0 slope in the S-Map procedure, both before and after self-learning.
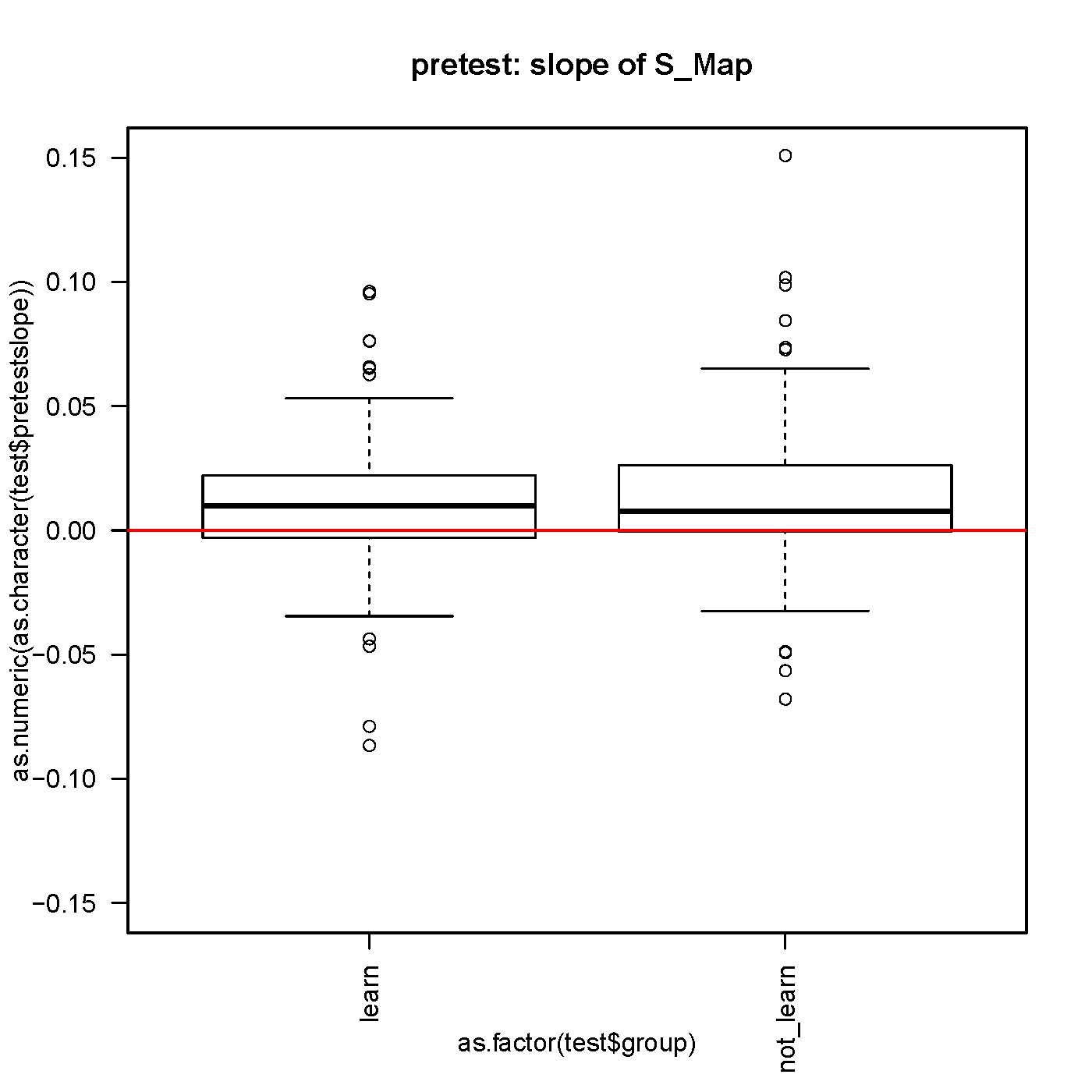
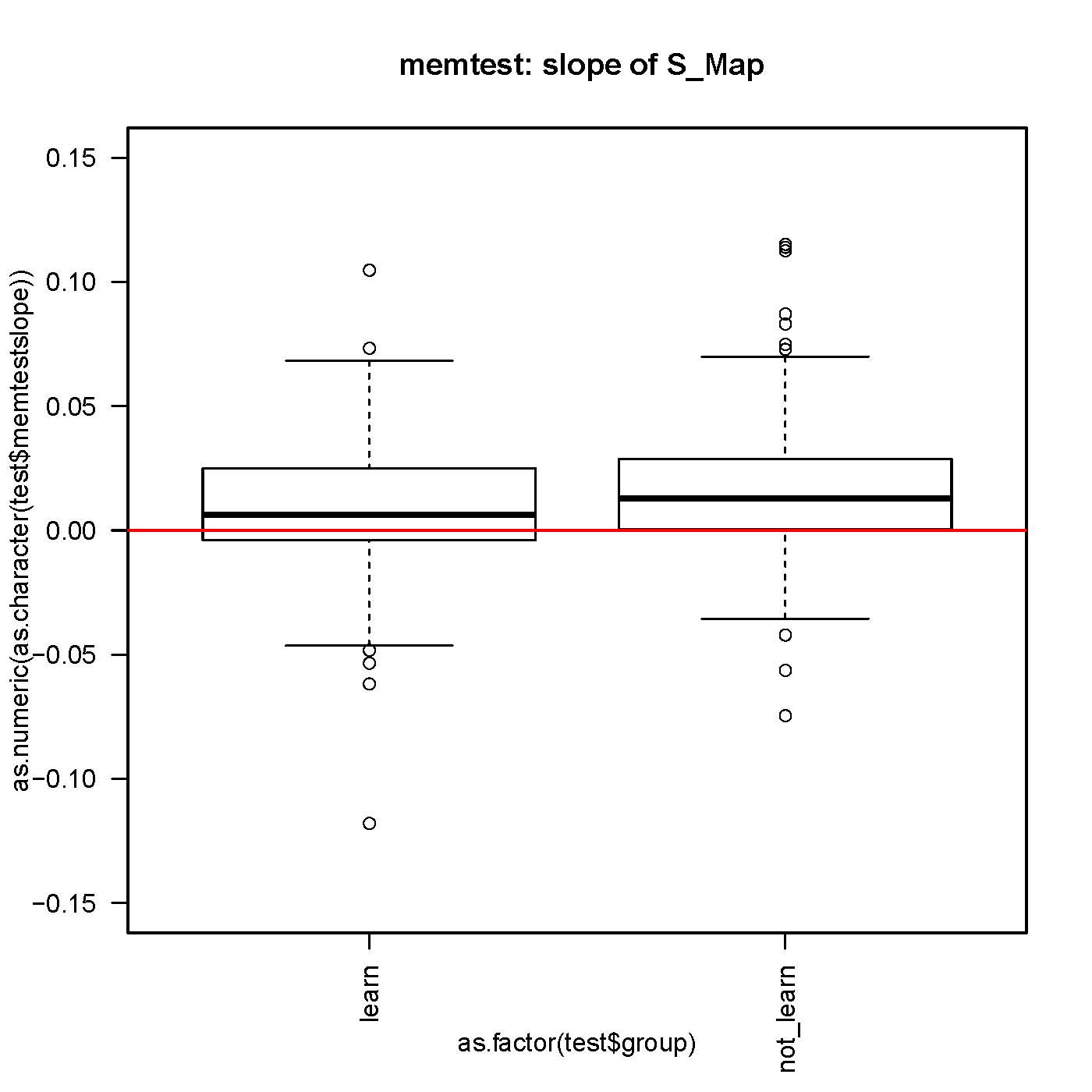
No difference in the slope while comparing before and after learning for each fly.
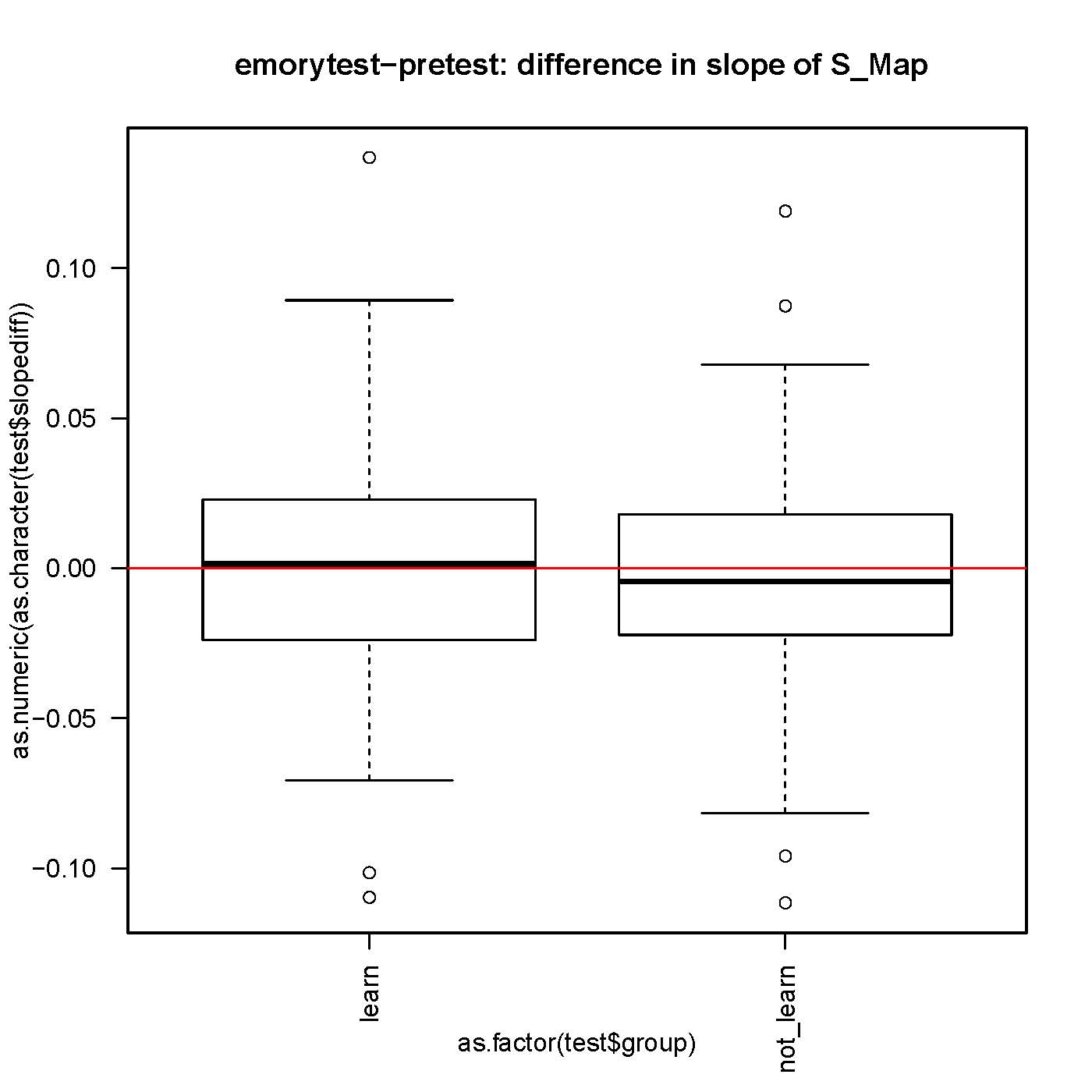
n>130 for each group.
FoxP anatomy
I had a look at the brain preparation I did last week (dicer2,FoxP RNAi x elavGal4 and controls, with nc82 antibody immunostaining). They look surprisingly good under the fluorescent microscope.
We still need to make confocal scans. I will ask Hilde whether she would be interested to do it…
PKC53E
I got mutant from Amita and will test them when out of quarantine. Then, we will have a clear answer about the compensatory mechanisms (the former mutant I tested was probably not null). There is also 2 other RNAi lines for PKC53e from the TriP project.
I am waiting for chrisi to be back and we will see what flies we need to order to bloomington in addition to these two. (There is also another RNAi against FoxP…)
The outcross of the RNAi is also running (3d cross, now).
We will then have 3 RNAi and one mutant, if the results are consistent, this would be enough to convince people that PKC53e is the PKC involved.