Finally done: rut and rsh flies better at self-learning
on Monday, January 22nd, 2024 1:36 | by Björn Brembs
At long last, I got all the flies together that we need for sufficient statistical power. As the preliminary data had indicated, WTB flies don’t learn with the short training, while rut and rsh flies do just fine.
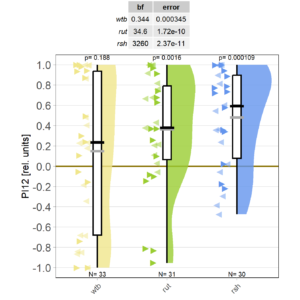
However, this may be due to genetic background effects, so we need to check the CRISPR mutants.
Category: Operant learning, operant self-learning, Radish | No Comments
Cloning via DNA Assembly
on Friday, September 1st, 2023 7:26 | by Isabel Stark
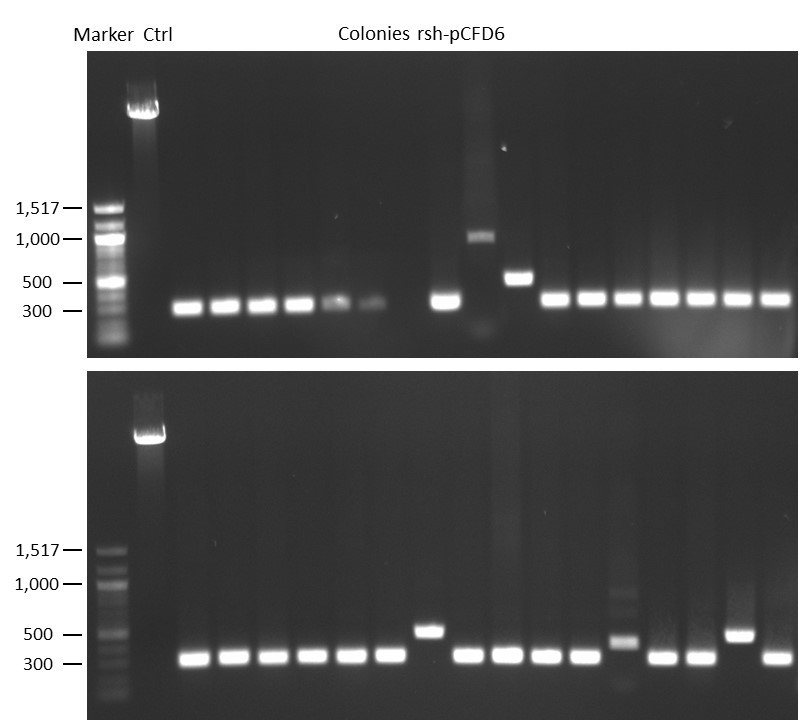
DNA Assembly in a 1:2 ratio of vector to insert with gRNAs of rsh and rut (Q5 and template concentration: 640 pg/µl) and 100ng of pCFD6 BbsI AP (using QuickCIP). Heat-shock (hs) transformation into E. coli (DH5α competent) with 10 µl Assembly Reaction and 100 µl cells.
For Crtl, pCFD6 BbsI AP was wrongly used.
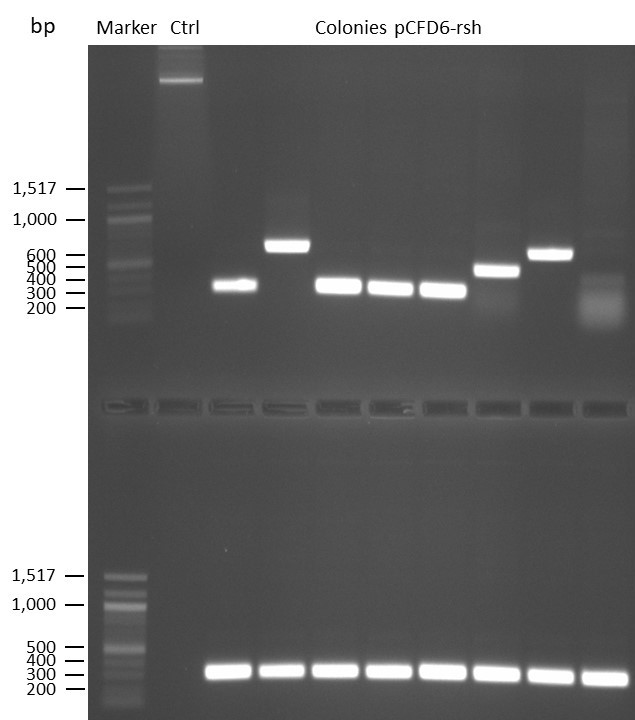
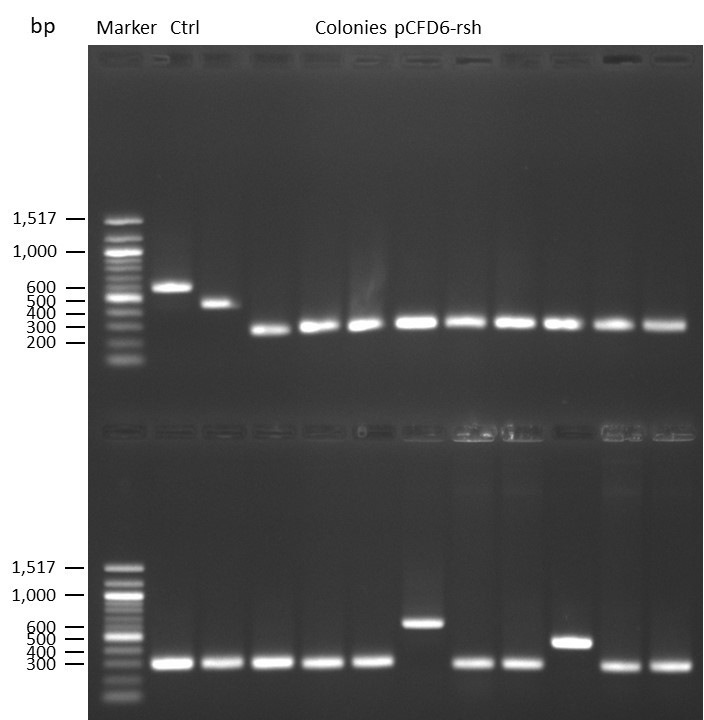
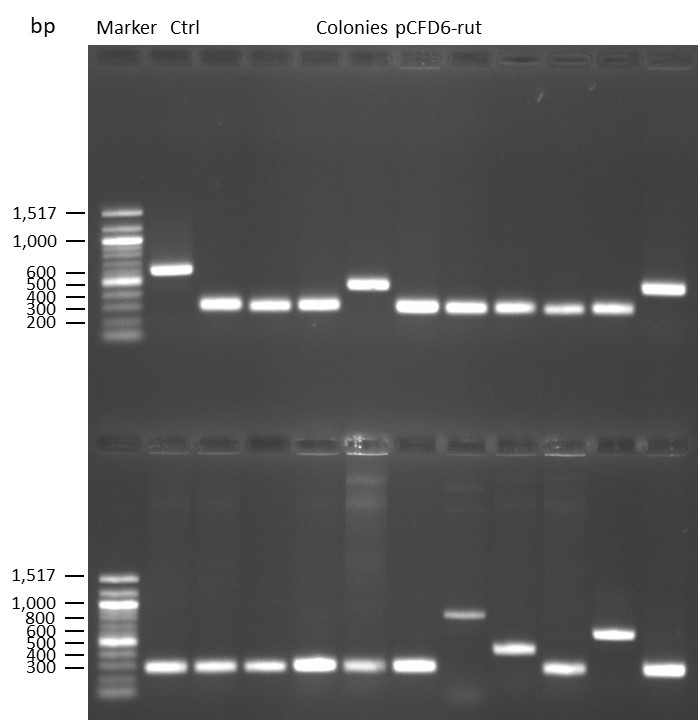
DNA Assembly in a 1:2 ratio of vector to insert with the gRNAs of rsh and rut (Q5 and template concentration: 640 pg/µl) and 100ng of pCFD6 BbsI AP (using FastAP and 2 extraction steps). Heat-shock (hs) transformation into E. coli (DH5α competent) with 10 µl Assembly Reaction and 100 µl cells.
For Crtl, pCFD6 BbsI AP was used in reaction.
Category: genetics, Memory, Operant learning, operant self-learning, Radish | No Comments
Success: rsh Stock has rsh1 Mutation
on Monday, August 7th, 2023 11:11 | by Isabel Stark
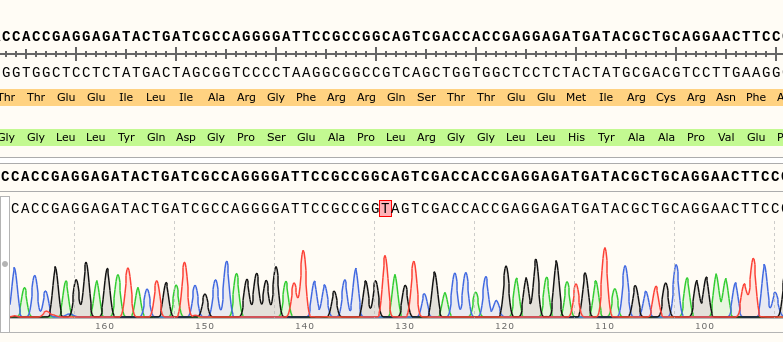
Via gDNA analysis and PCR was the specific area of the rsh gene extracted and amplified where the nucleotide substitution: C to T (Folkers et al., 2006) should be for the rsh1 mutation. The amplicon was Sanger sequenced which proved the nucleotide substitution.
Category: genetics, Memory, Operant learning, operant self-learning, Radish, Uncategorized | No Comments
Little by little
on Monday, July 24th, 2023 1:59 | by Björn Brembs
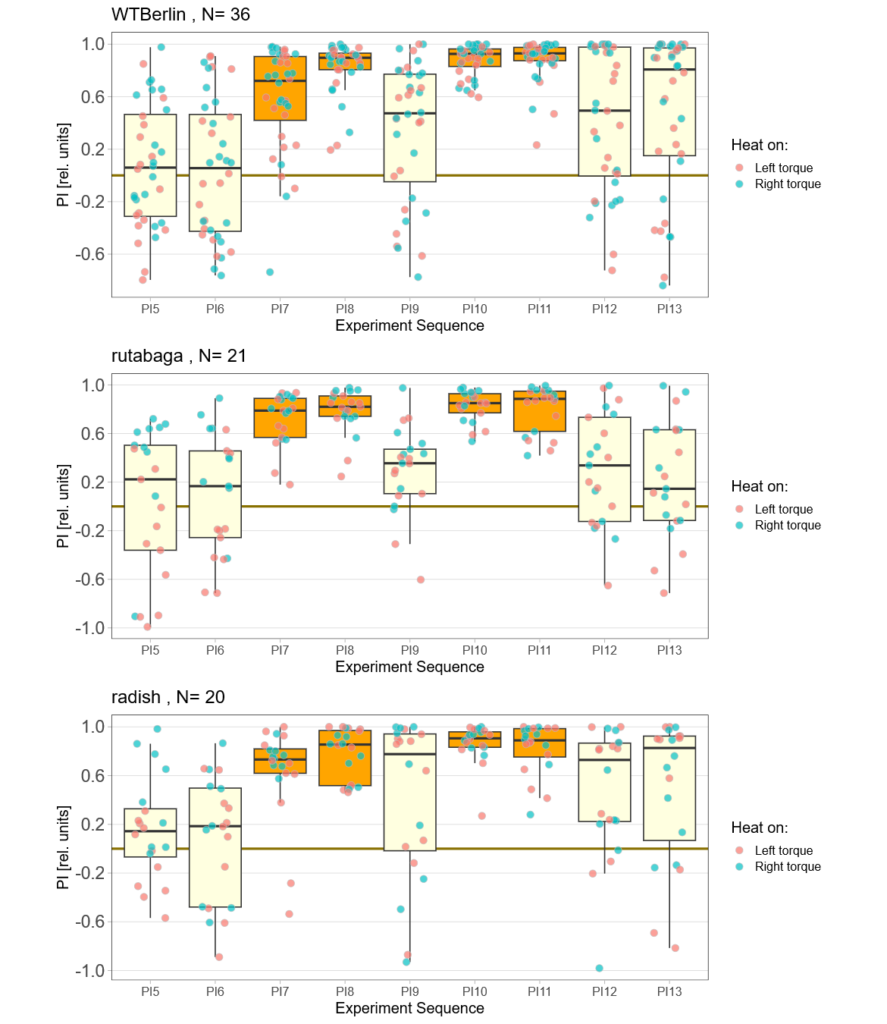
Despite very warm weather, some flies did fly, even though the learning performance of the control flies was really poor. At least for now, it looks like all stocks are learning and that rut and rsh flies learn at least equally well as the Berlin flies. I’ve also managed to fix the positive preference problem:
Category: operant self-learning, Radish | No Comments
Creating gRNAs via PCR
on Friday, July 7th, 2023 6:38 | by Isabel Stark
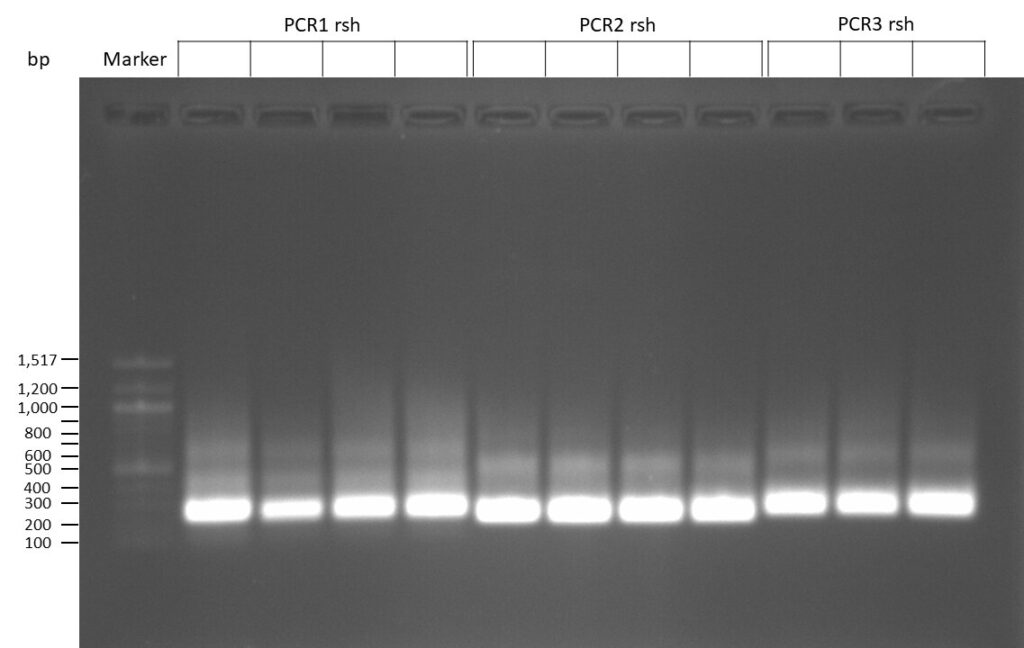
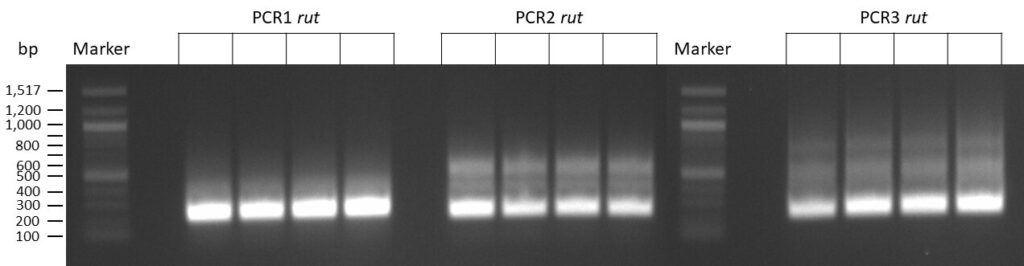
The template pCFD6 was used with a concentration of 640 pg/µl.
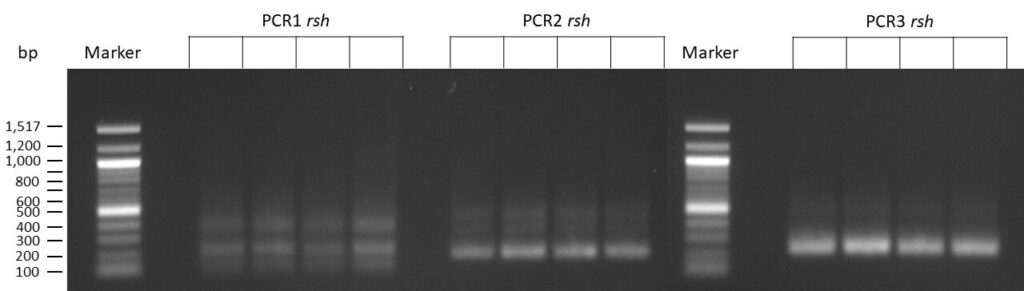
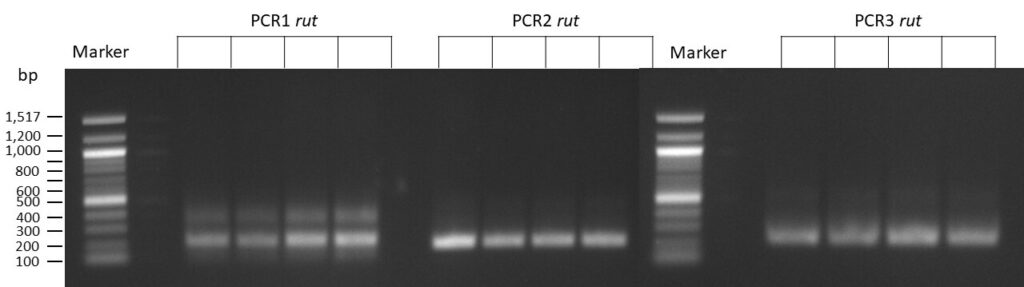
The template pCFD6 was used with a concentration of 64 pg/µl (1:10 dilution).
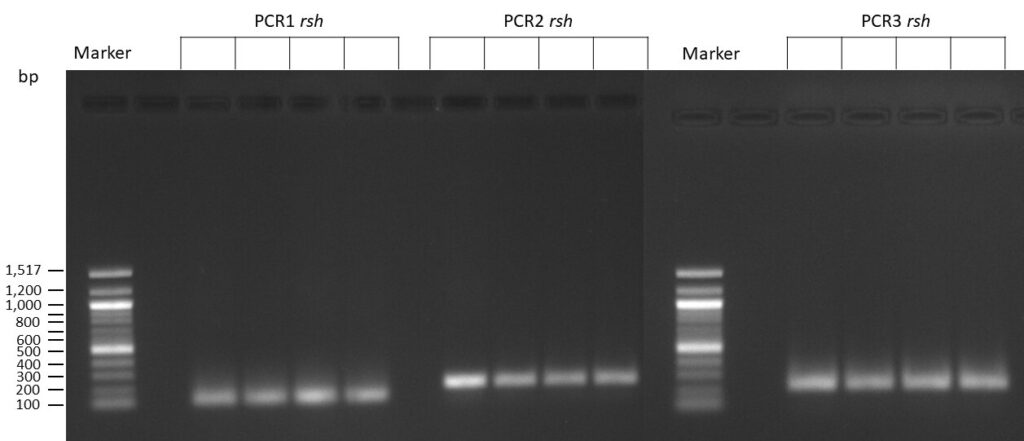
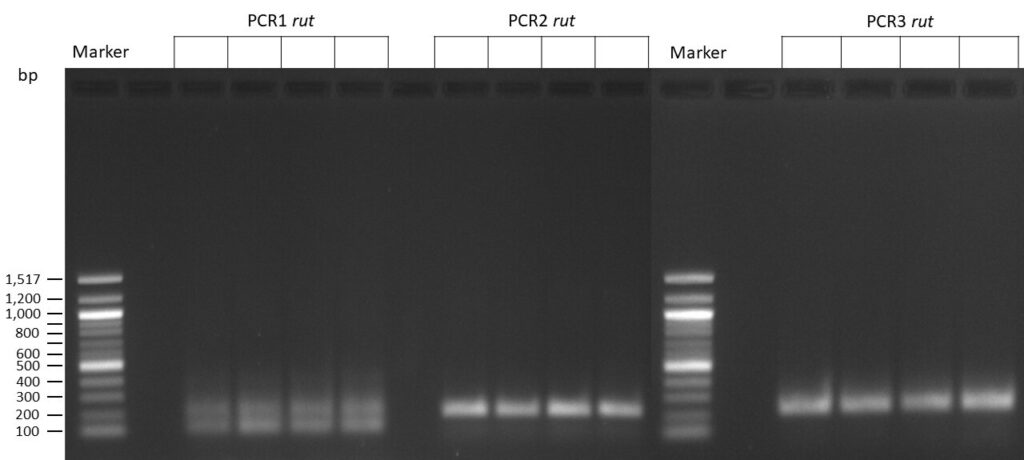
The template pCFD6 was used with a concentration of 64 pg/µl (1:10 dilution).
50µl of 5xQ5 High GC Enhancer was added to the PCR mix.
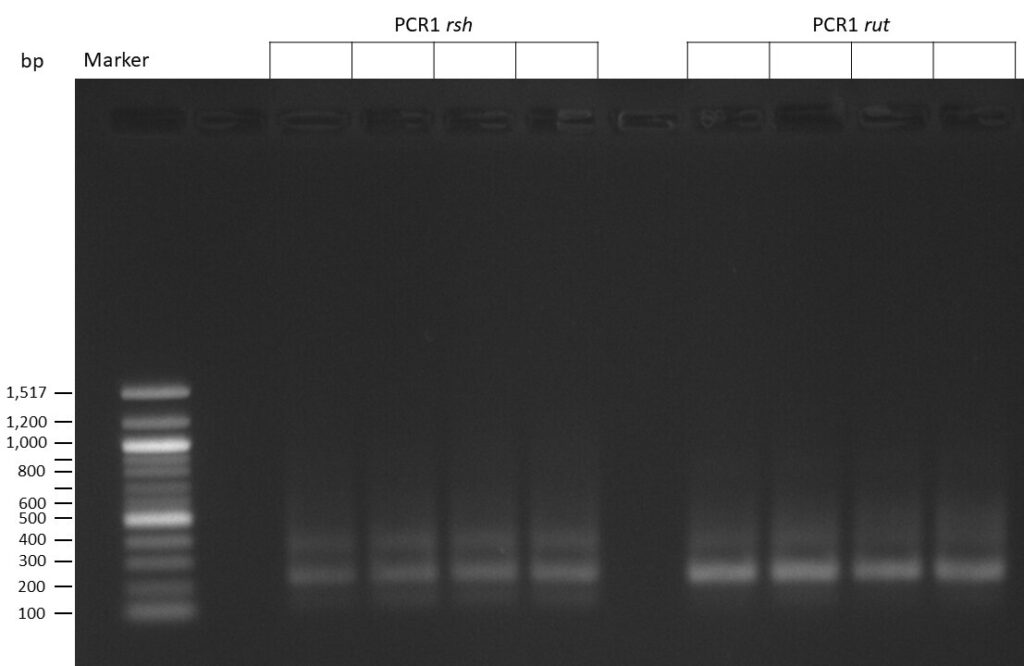
The template pCFD6 was used with a concentration of 128 pg/µl (1:5 dilution).
50µl of 5xQ5 High GC Enhancer was added to the PCR mix.
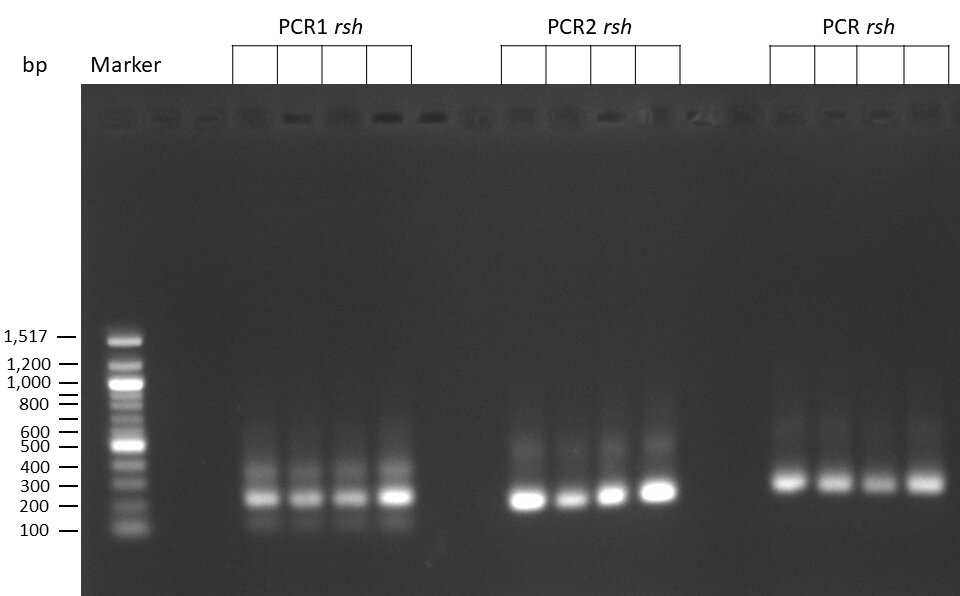
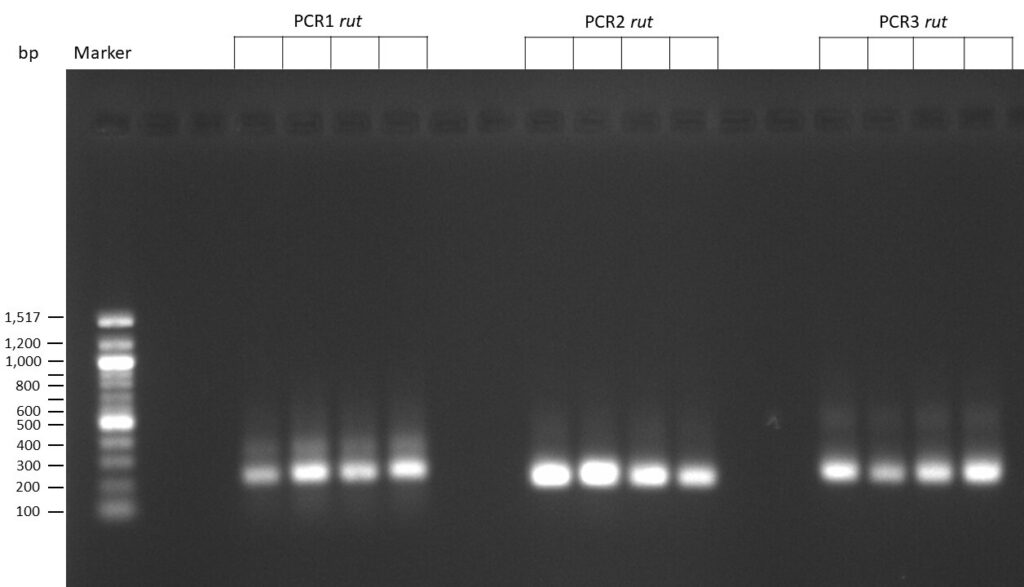
The template pCFD6 was used with a concentration of 640 pg/µl.
The Phusion DNA Polymerase was used instead of the Q5 High-Fidelity DNA Polymerase.
Category: genetics, Memory, Operant learning, operant self-learning, Radish, Uncategorized | No Comments
