Success: rsh Stock has rsh1 Mutation
on Monday, August 7th, 2023 11:11 | by Isabel Stark
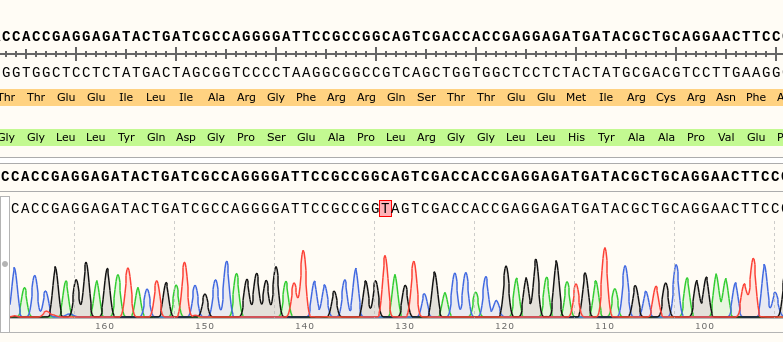
Via gDNA analysis and PCR was the specific area of the rsh gene extracted and amplified where the nucleotide substitution: C to T (Folkers et al., 2006) should be for the rsh1 mutation. The amplicon was Sanger sequenced which proved the nucleotide substitution.
Category: genetics, Memory, Operant learning, operant self-learning, Radish, Uncategorized | No Comments
Creating gRNAs via PCR
on Friday, July 7th, 2023 6:38 | by Isabel Stark
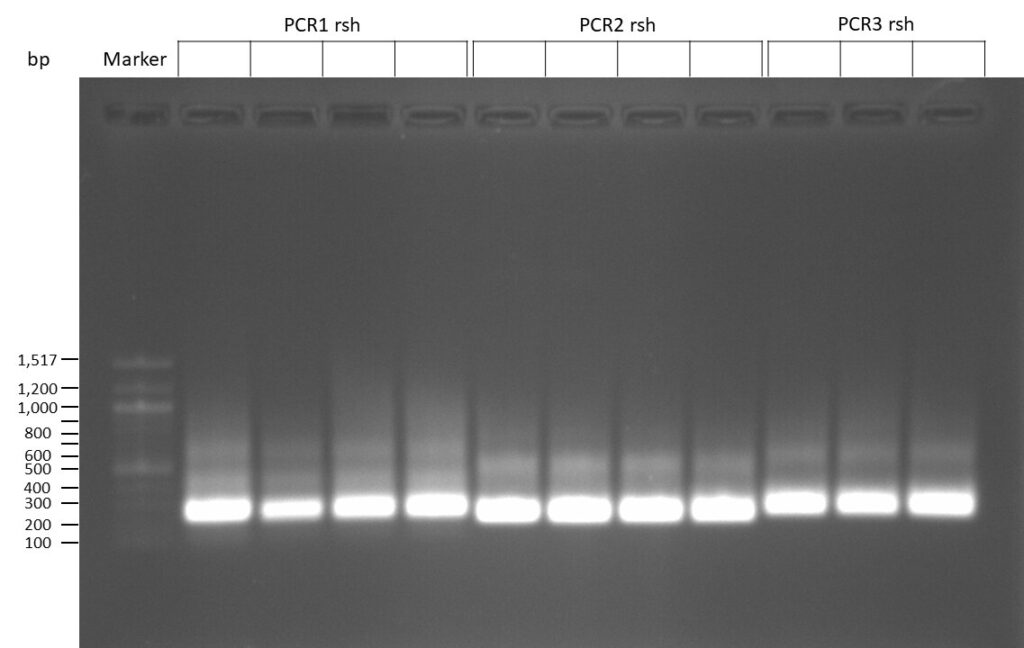
The template pCFD6 was used with a concentration of 640 pg/µl.
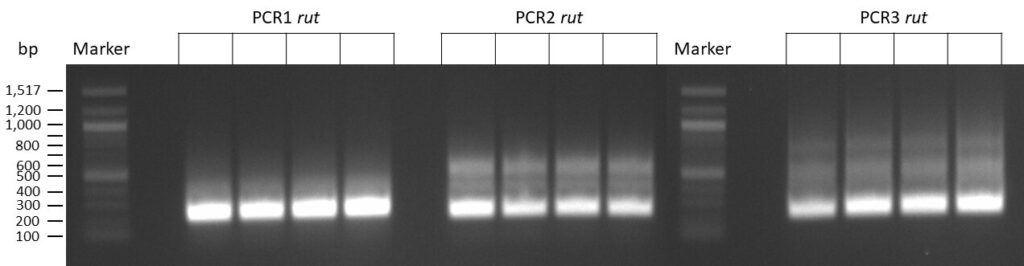
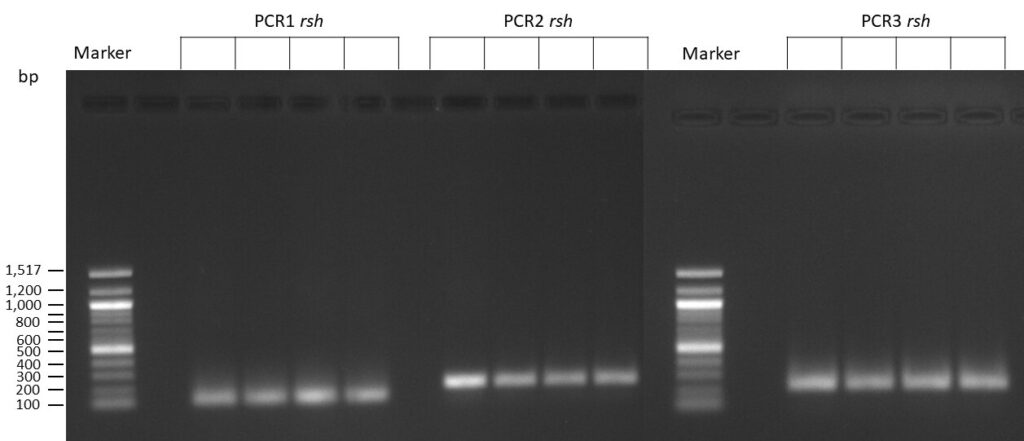
The template pCFD6 was used with a concentration of 64 pg/µl (1:10 dilution).
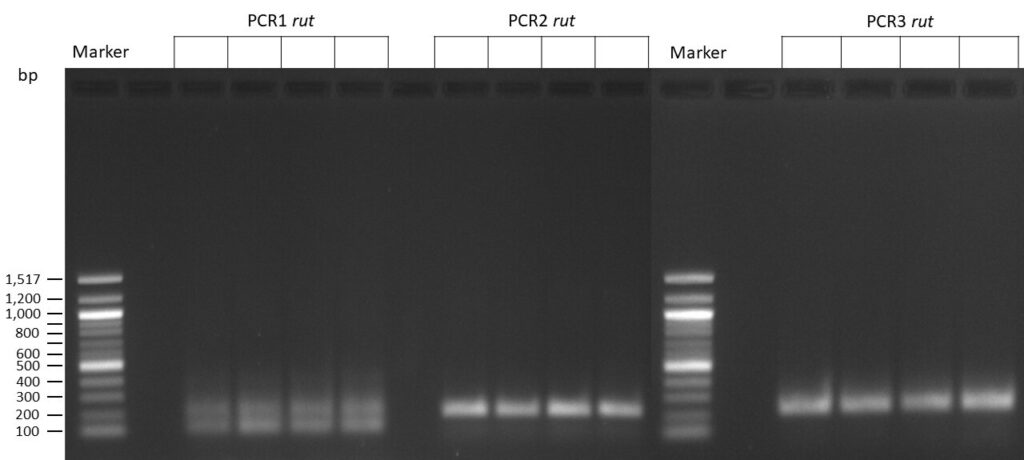
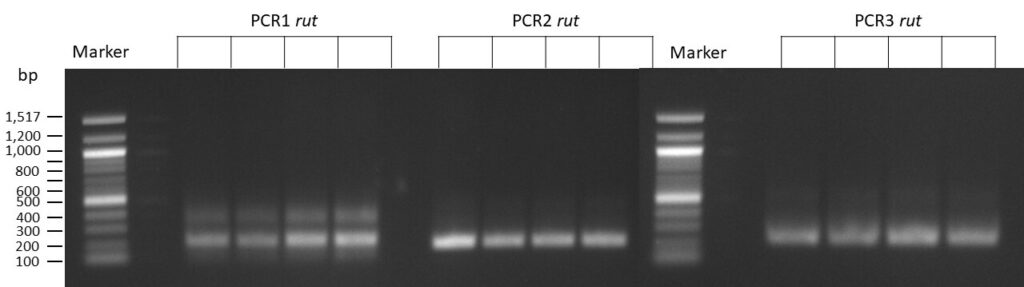
The template pCFD6 was used with a concentration of 64 pg/µl (1:10 dilution).
50µl of 5xQ5 High GC Enhancer was added to the PCR mix.
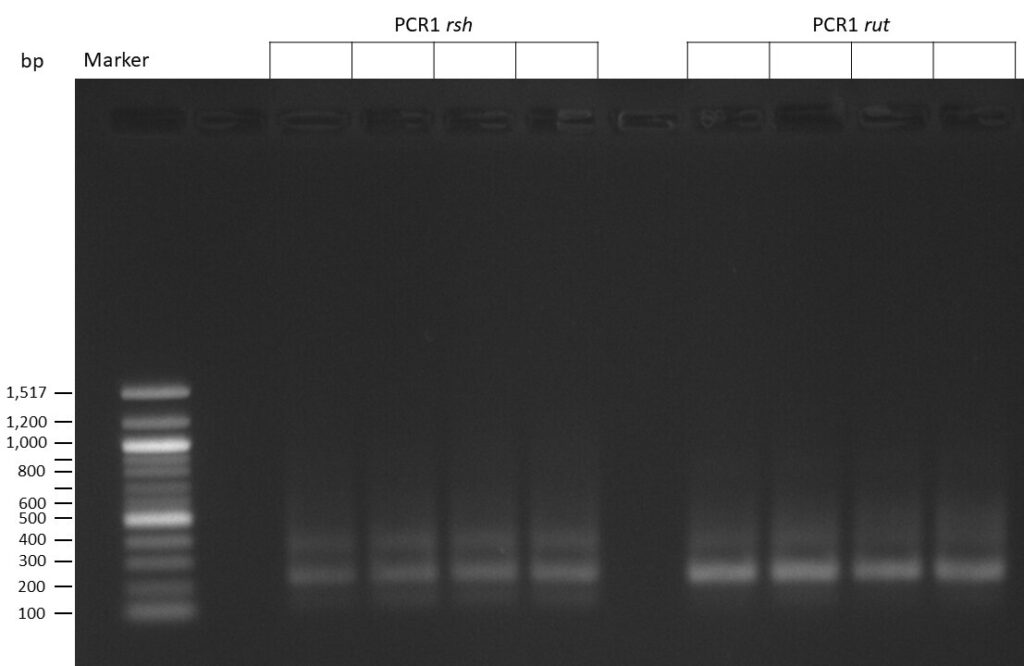
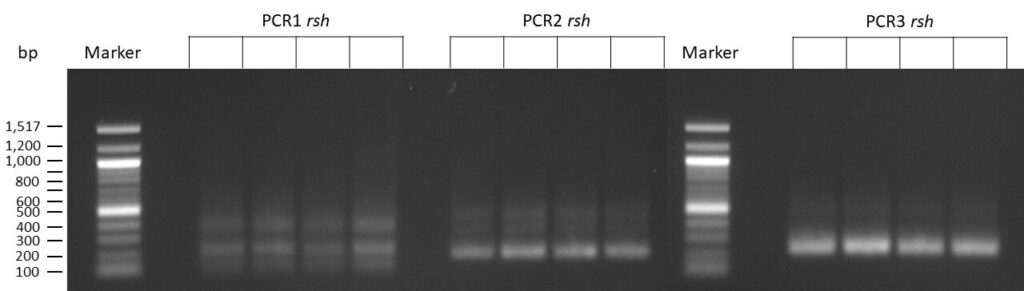
The template pCFD6 was used with a concentration of 128 pg/µl (1:5 dilution).
50µl of 5xQ5 High GC Enhancer was added to the PCR mix.
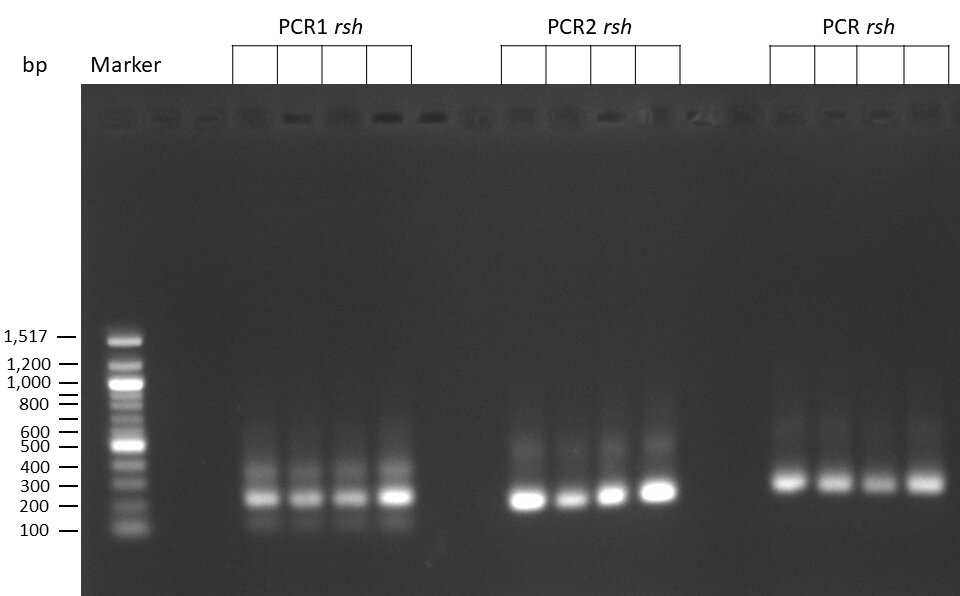
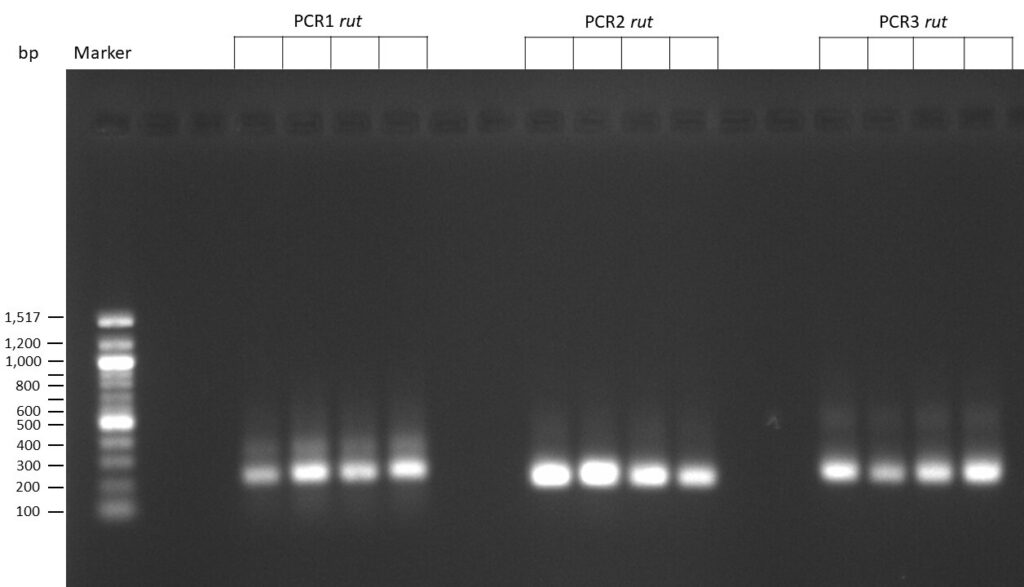
The template pCFD6 was used with a concentration of 640 pg/µl.
The Phusion DNA Polymerase was used instead of the Q5 High-Fidelity DNA Polymerase.
Category: genetics, Memory, Operant learning, operant self-learning, Radish, Uncategorized | No Comments