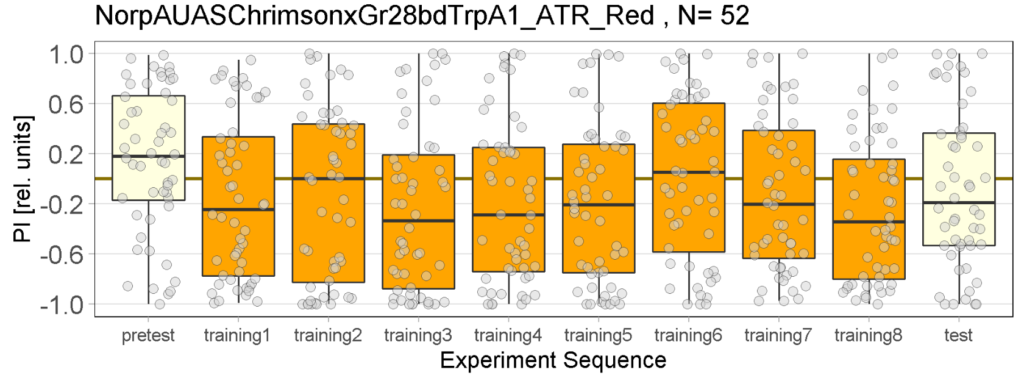
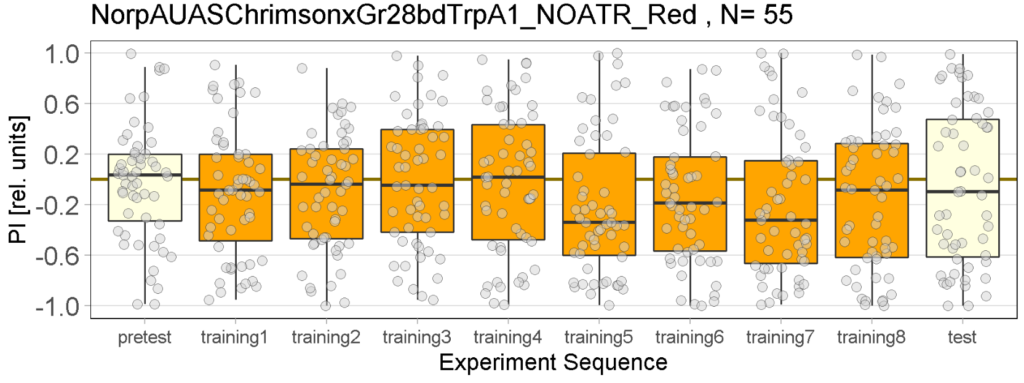
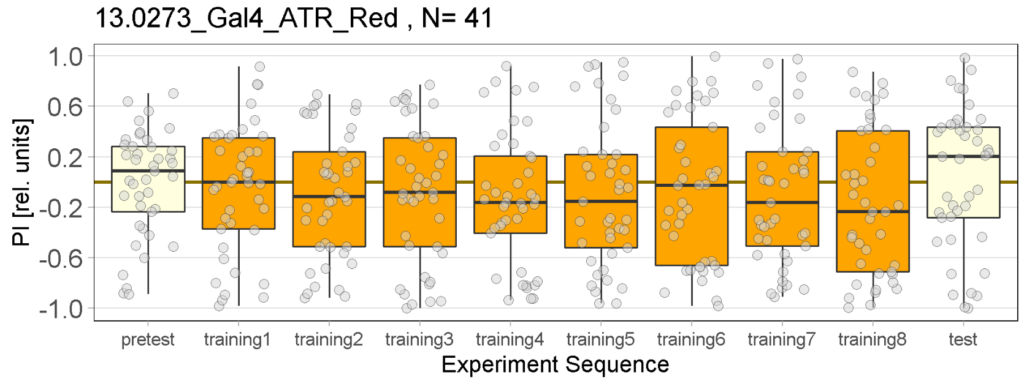
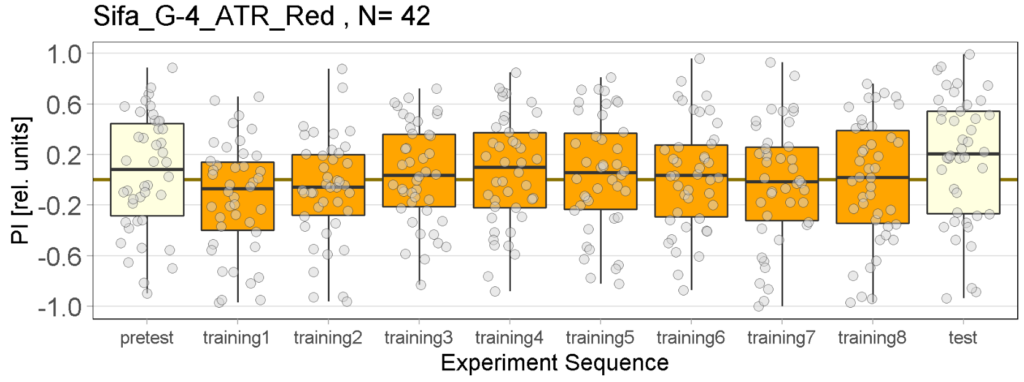
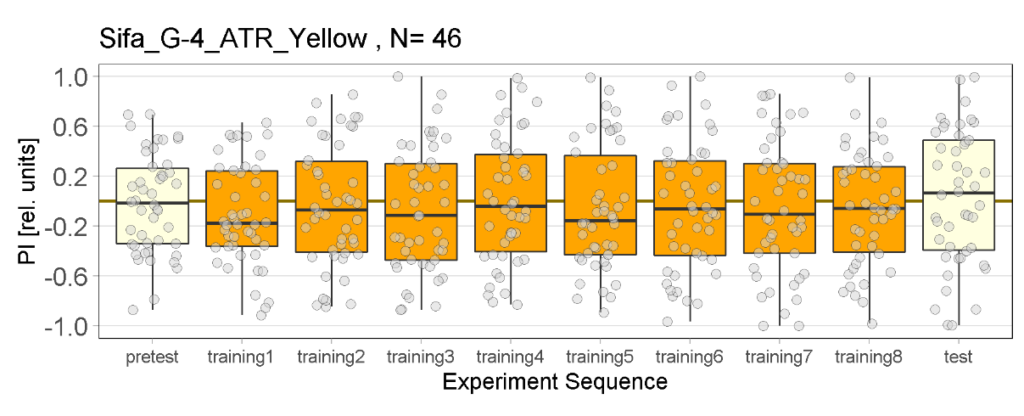
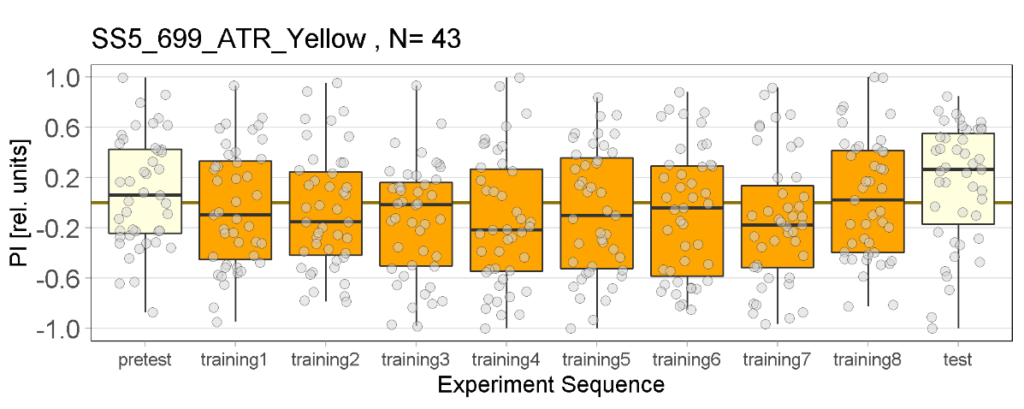
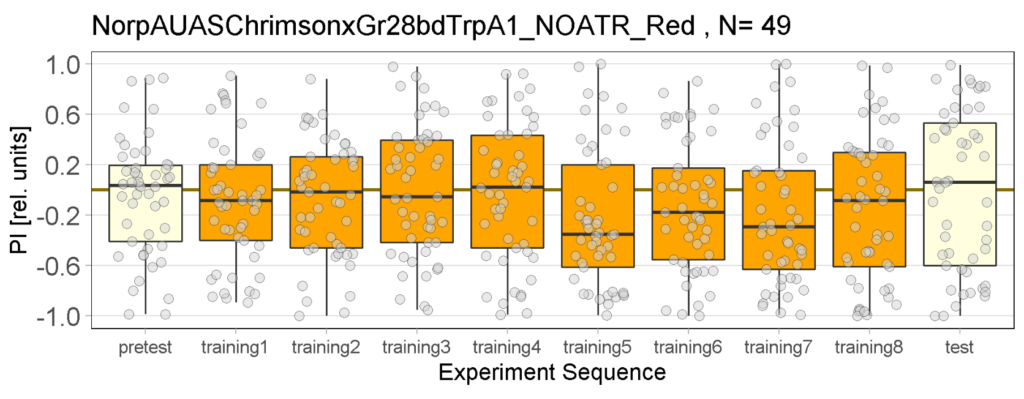
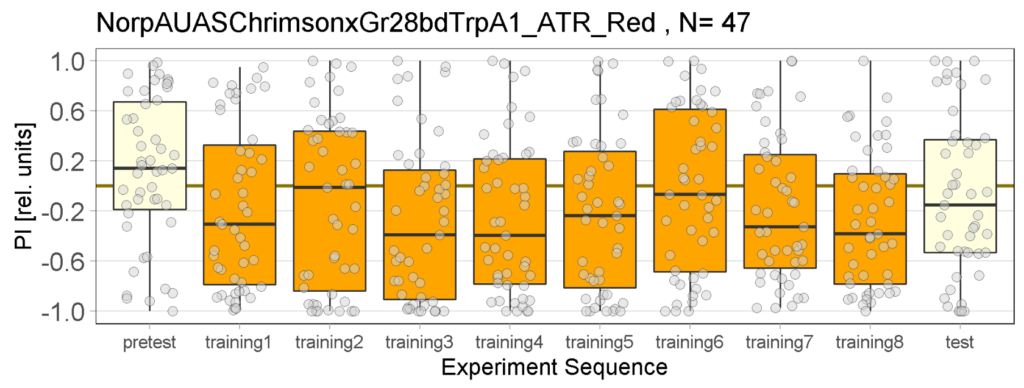
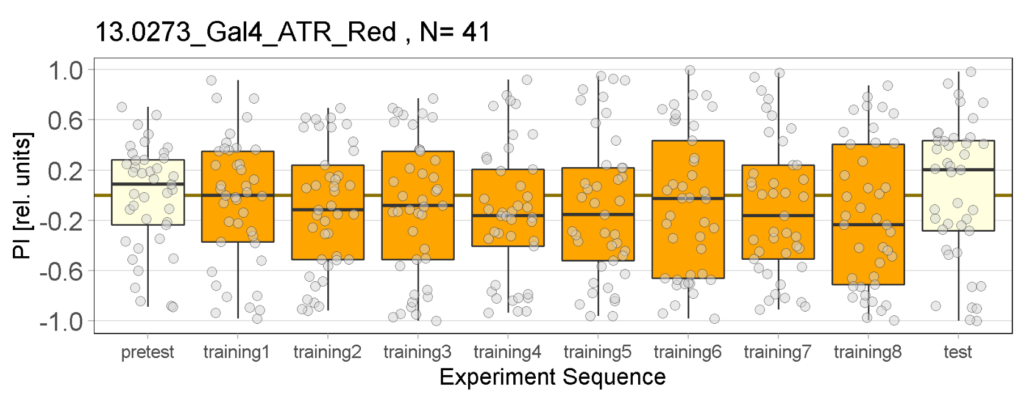
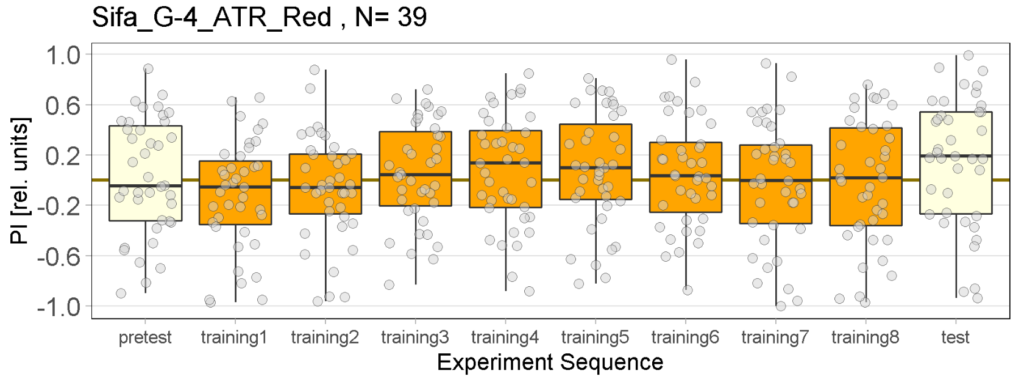
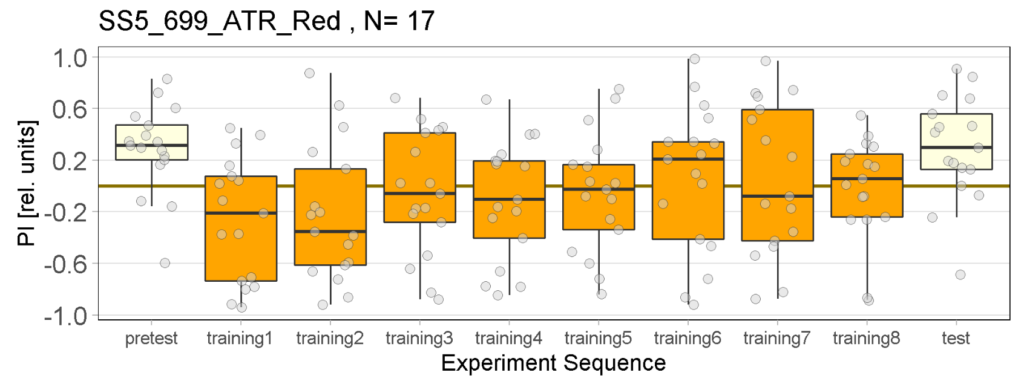
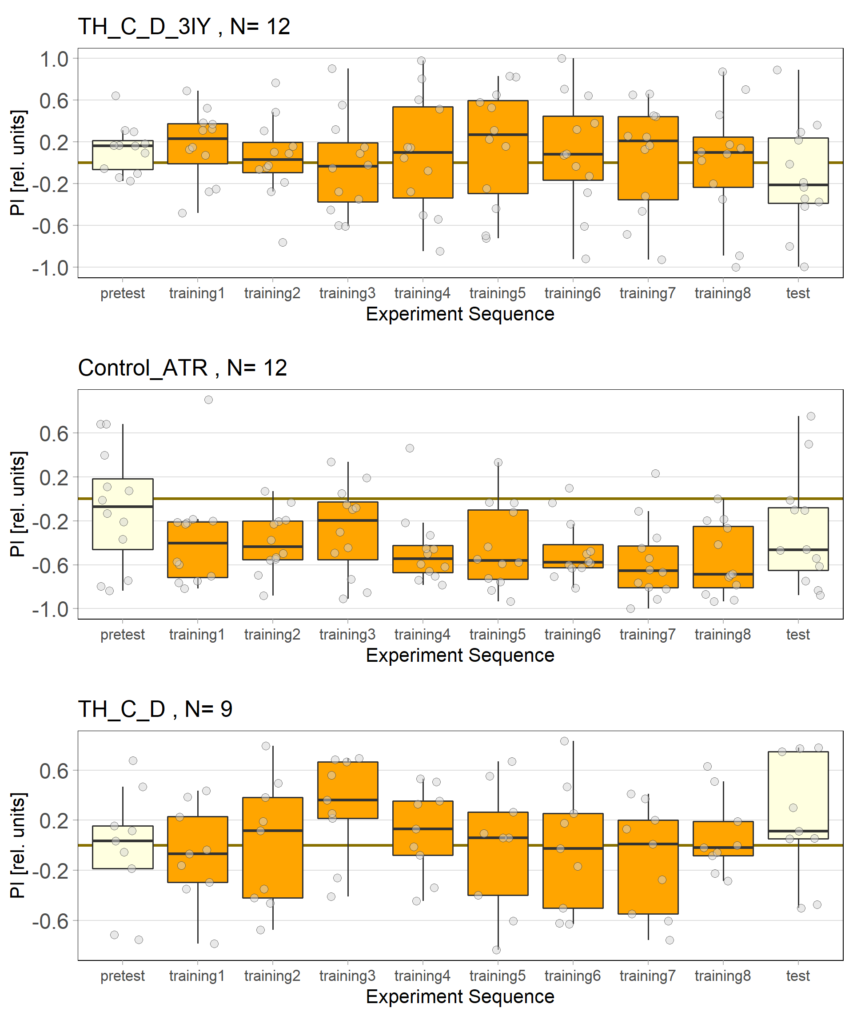
Joystick Results for 2-minute-testing
on Tuesday, May 30th, 2023 1:31 | by Luisa Guyton
The following figures show the results of the joystick test using the same test line as before. In addition, the flies were fed not only ATR but also 3IY (3-iodo-L-tyrosine), an inhibitor of dopamine synthesis. This was done to determine whether the previously observed effect was due to dopamine alone and could therefore be suppressed by the inhibitor, or whether other factors also influenced the flies’ behaviour in the joystick test.
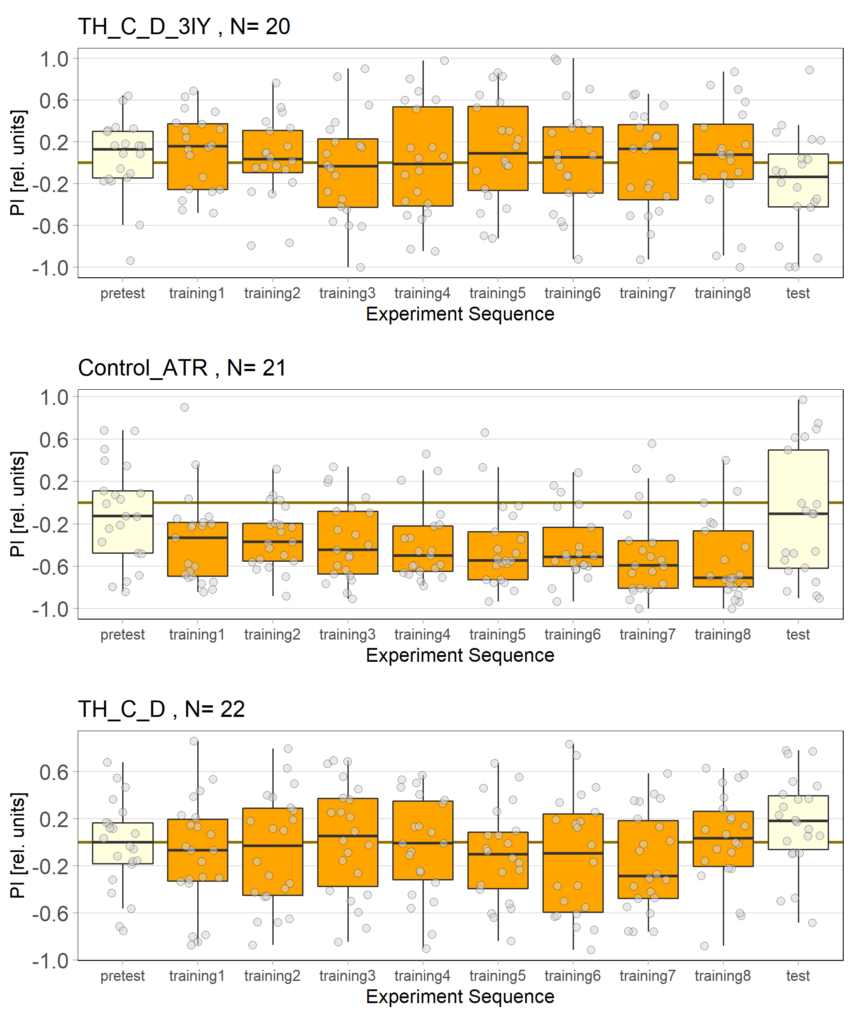
Category: Optogenetics | No Comments
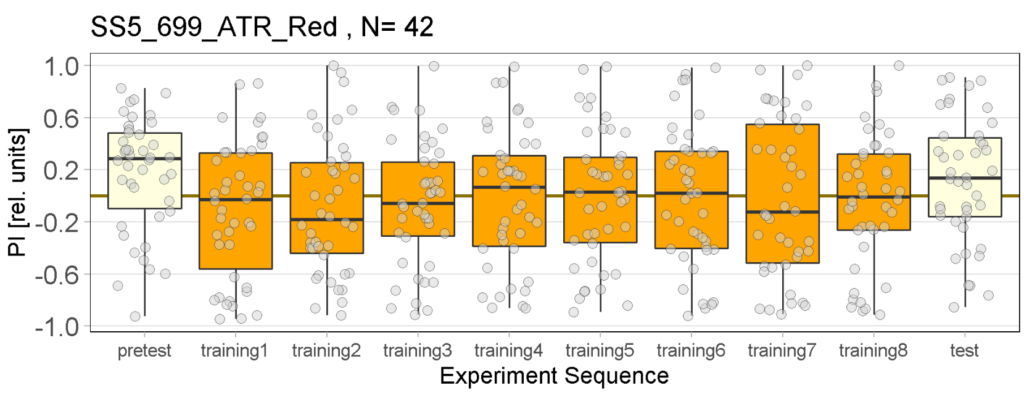
Red light Joystick Results
on Monday, April 3rd, 2023 12:33 | by Luisa Guyton
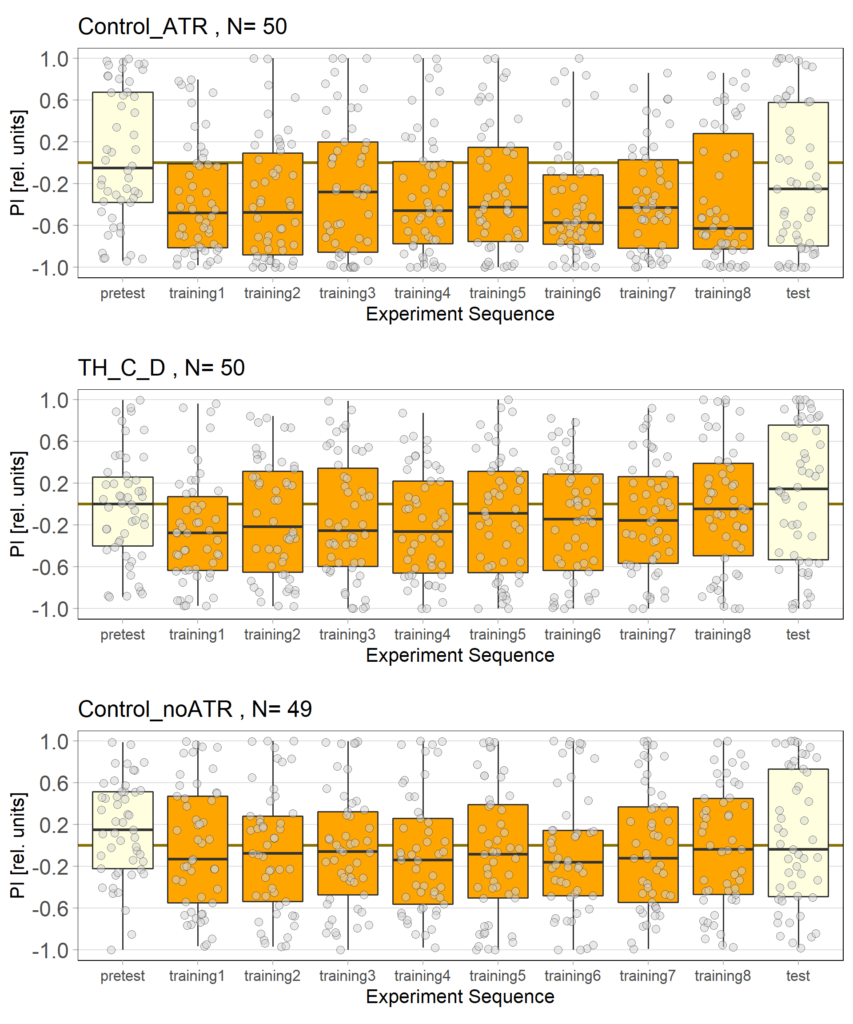
Category: Optogenetics | No Comments
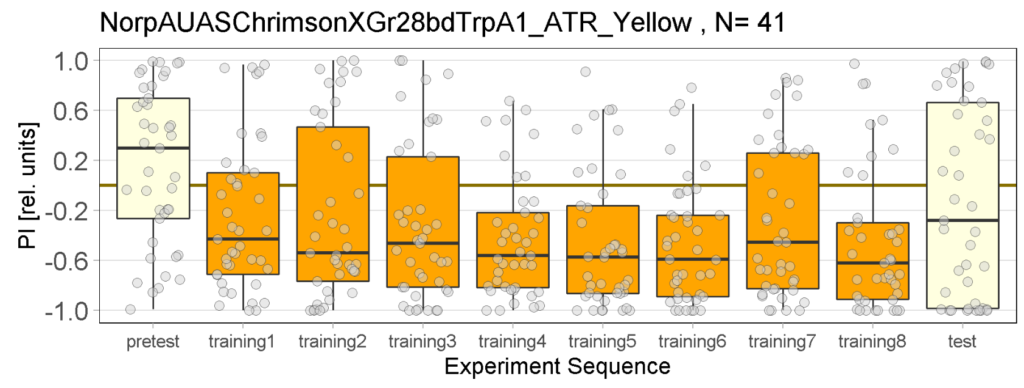
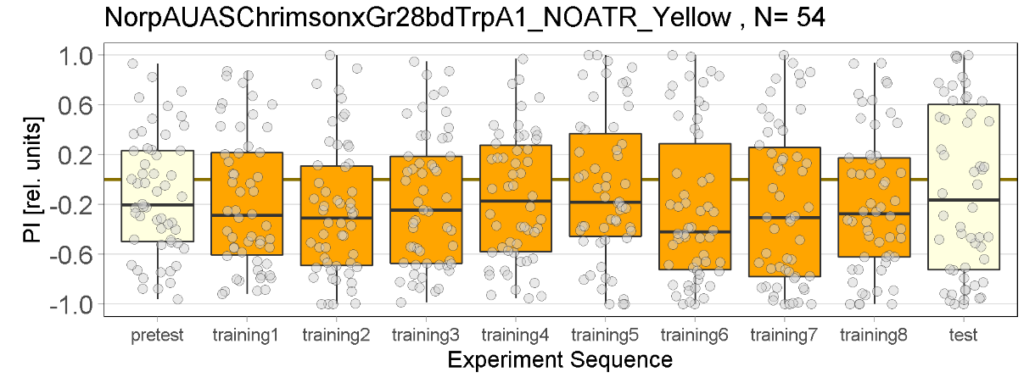
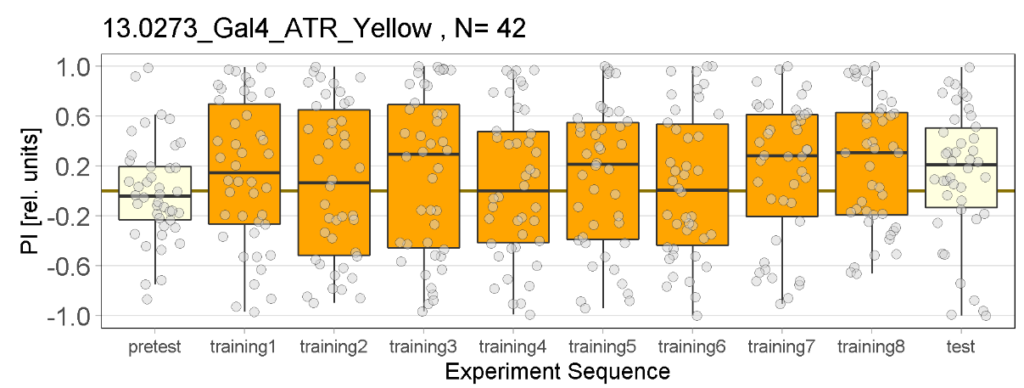
Yellow light Joystick Results
on Monday, March 27th, 2023 11:57 | by Luisa Guyton

Category: Optogenetics | No Comments
Update: Red light Joystick results
on Saturday, March 18th, 2023 1:28 | by Florian Hierstetter
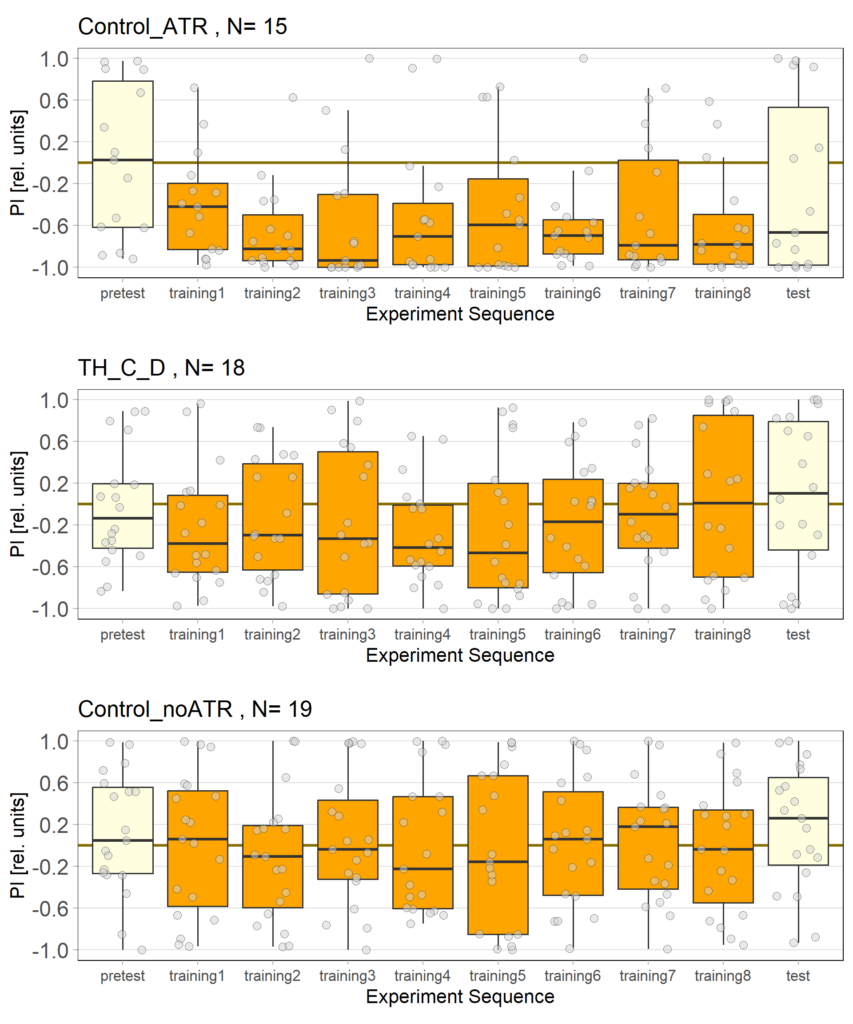
After the first full week of testing the three different fly lines with red light, we have analyzed the results. We are now able to provide a further update on our findings.
While the effect for the control groups seems to stay consistent with our previous findings, the effect for the positive control has weakened and is closer to the strength measured in previous experiments. We utilized the norpAP24;Gr28bd+TrpA1>Chrimson line for both our positive and negative controls.
Furthermore, the experimental group, which previously favored red light in the post-test, showed a more varied and inconclusive profile. We will need to conduct further testing to determine whether the previously observed effect was simply caused by the small sample size.
Another important discovery was the necessity of feeding the flies sugar water 5-10 minutes before the experiment in order to maintain their activity levels. This way the flies will be identified as a valid data point by the R-script and will ensure more and more accurate data. It is important to note that we will implement this feeding process in all future experiments to ensure consistency and accuracy of the data collected.
Category: Optogenetics | No Comments
Red light Joystick Results
on Tuesday, March 14th, 2023 8:57 | by Luisa Guyton
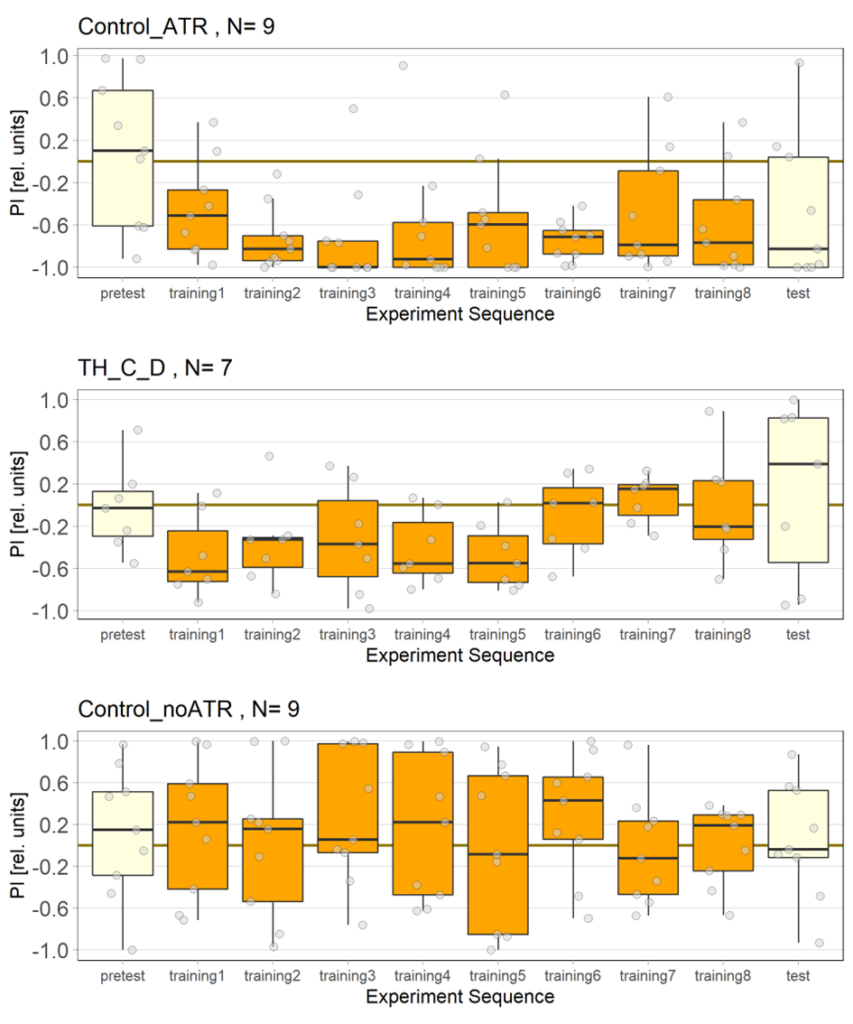
The light used is red and has an intensity of 400 to 470 lux, the LEDs have a voltage of 7.5 volts. The control flies observed seem to show a stronger effect than in previous tests, which could be due to the use of a thinner fishing line (0.6 mm), which is easier to place correctly in the clamp of the joystick machine.
For the negative and positive control we used the line norpAP24;Gr28bd+TrpA1>Chrimson.
Category: Optogenetics | No Comments
Yellow light_Comparison of final results in T-maze vs Joystick
on Wednesday, September 28th, 2022 2:22 | by Vivi Samara
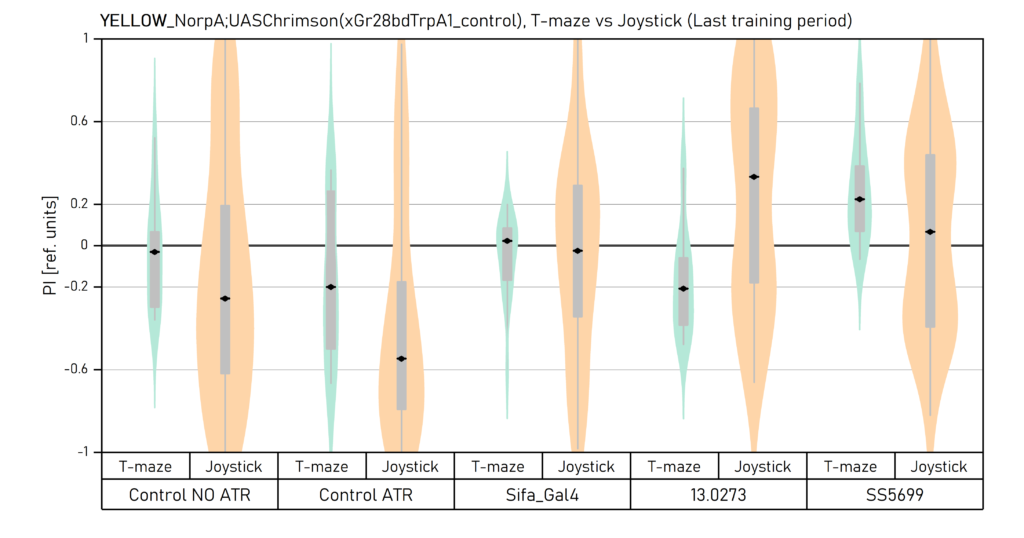
Final results from all the experimernts with yellow light. Data obtained by Aslihan, Enes and Vivi.
The population of each plot can be seen in the previous uploaded graphs.
Category: Optogenetics | No Comments
Yellow light final T-maze results
on Tuesday, September 27th, 2022 9:29 | by Vivi Samara
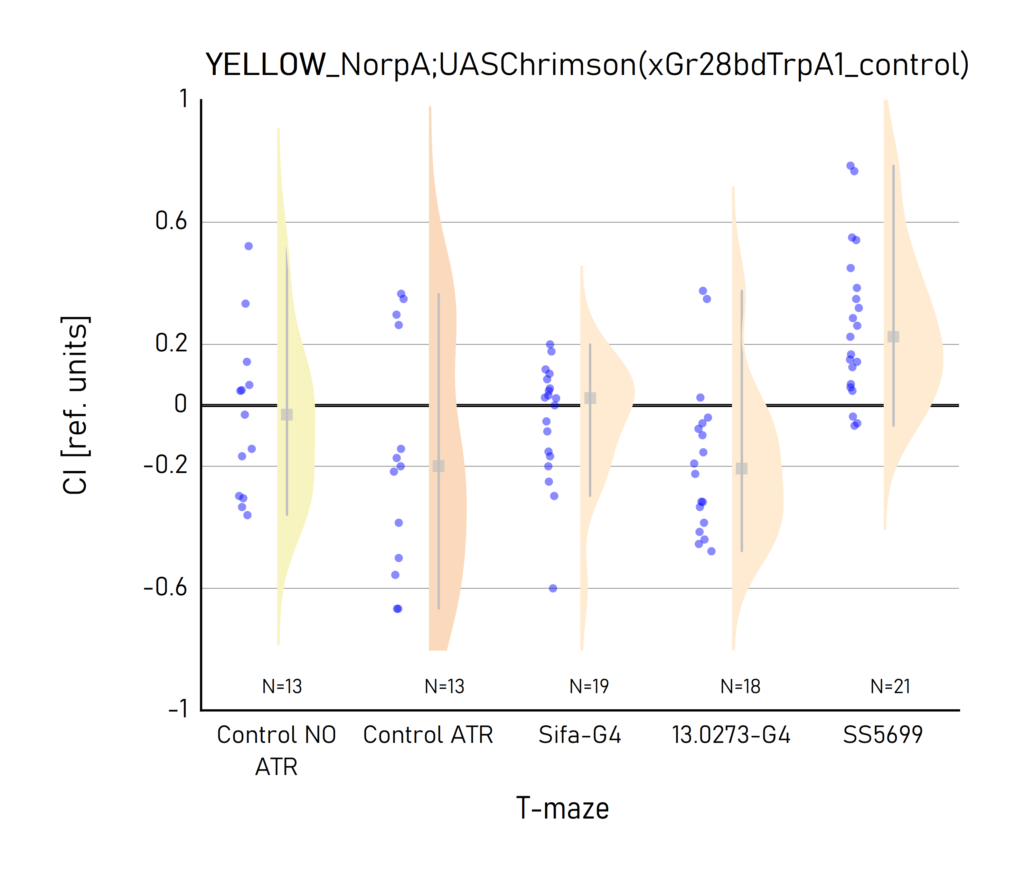
Results by Aslihan and Vivi.
Category: Optogenetics | No Comments
Red Joystick Results
on Friday, September 23rd, 2022 6:55 | by Enes Seker
Category: Optogenetics | No Comments
Yellow Joystick Results
on Friday, September 23rd, 2022 6:50 | by Enes Seker
Category: Optogenetics | No Comments
Red Joystick Results
on Monday, September 19th, 2022 9:40 | by Enes Seker
Category: Optogenetics | No Comments