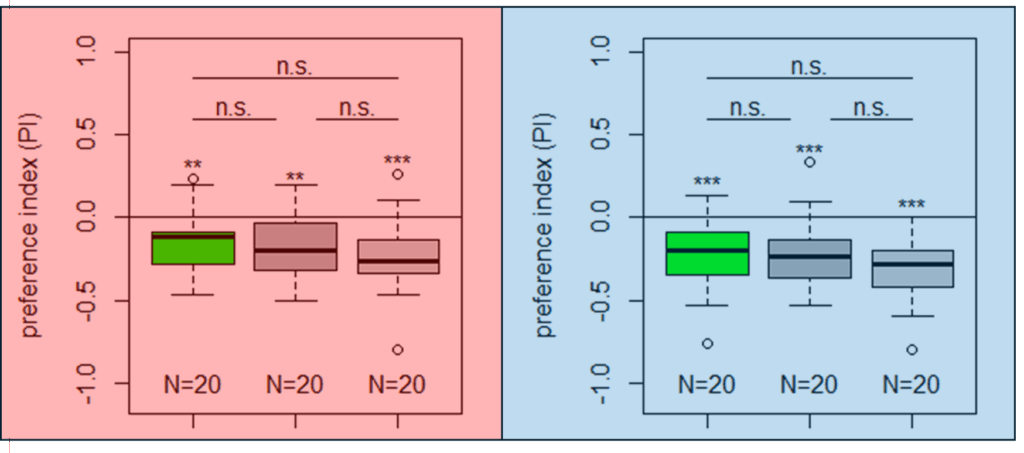
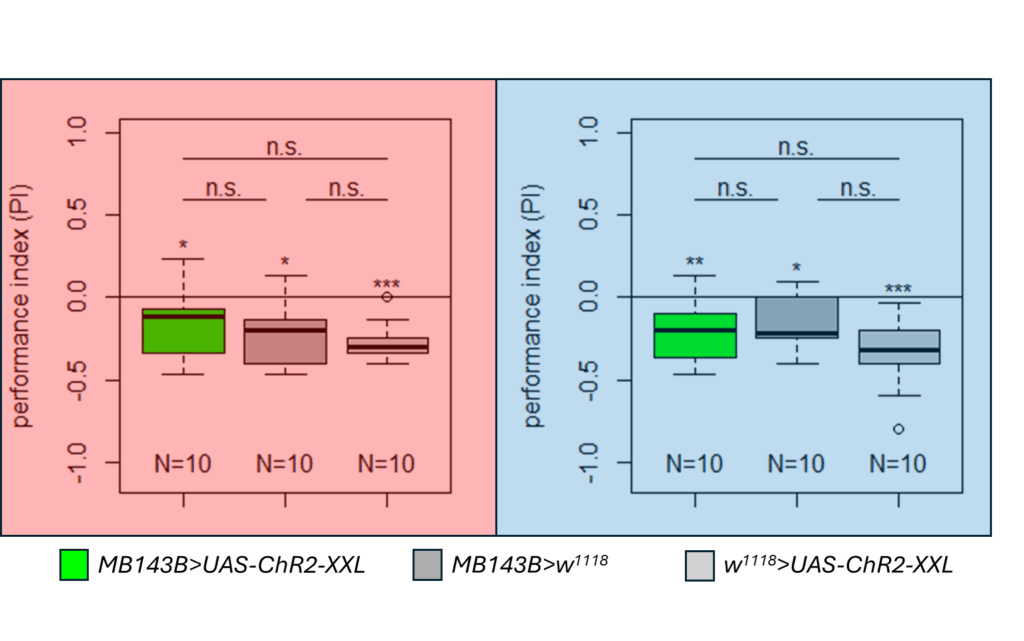
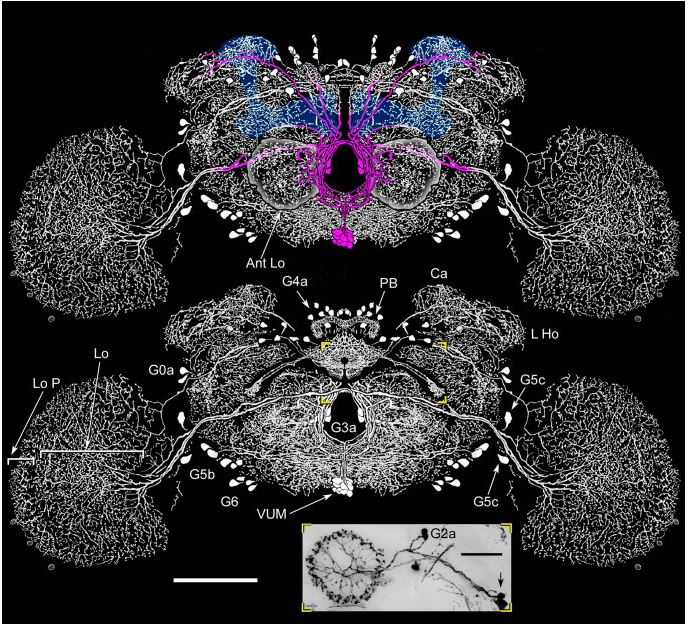
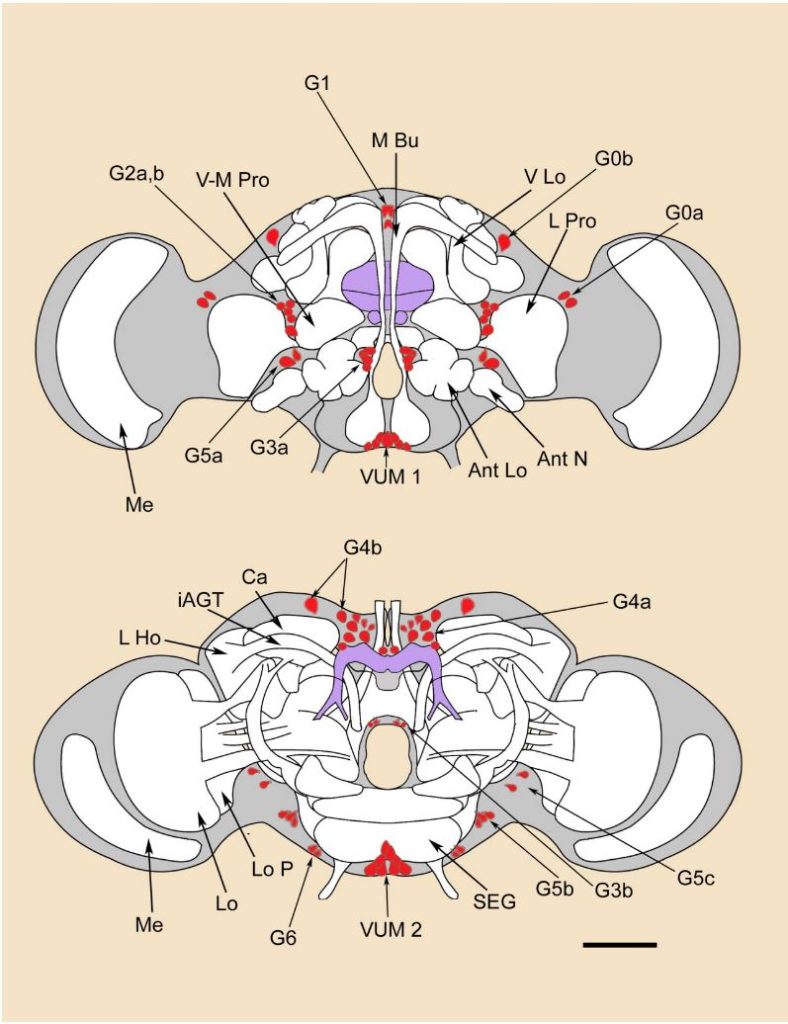
Yellow T-Maze Results
on Friday, November 14th, 2025 3:35 | by Daniel Döringer
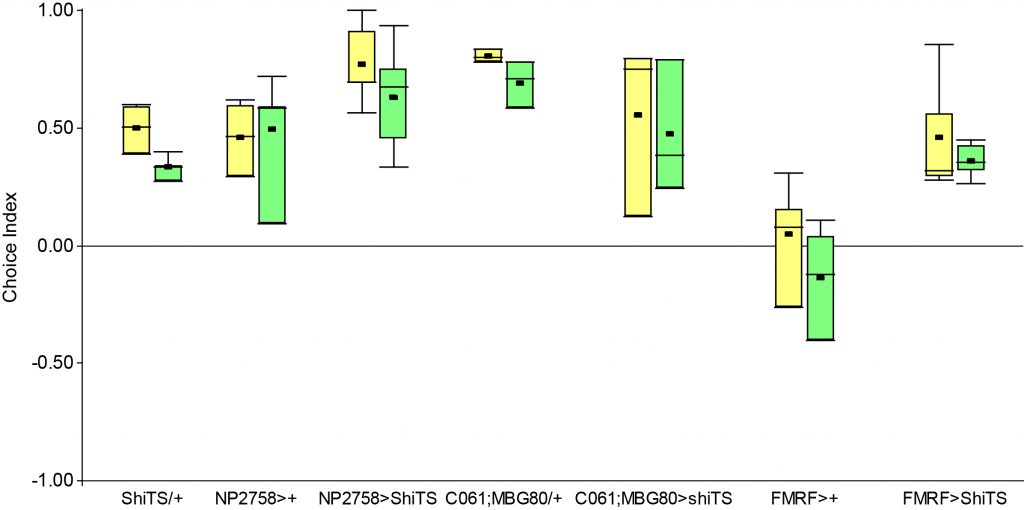
I performed the first set of T-Maze experiments, which included 3IY treated flies, with red light. In this experiment I could nicely reproduce earlier results in PPM2 flies only treated with ATR. Flies initially showed weak avoidance of optogenetic stimulation, and developed a weak approach-behavior over the time course of ten minutes. In the 3IY treated flies I found similar avoidance after 1 minute of testing but, interestingly, flies kept the same level of avoidance also for a choice time of 10 minutes. This indicates that initial avoidance might be independent of dopamine, but prolonged or repeated release of the neurotransmitter from PPM2 neurons might lead to circuit changes, weakening avoidance behavior, potentially even changing it to approach.
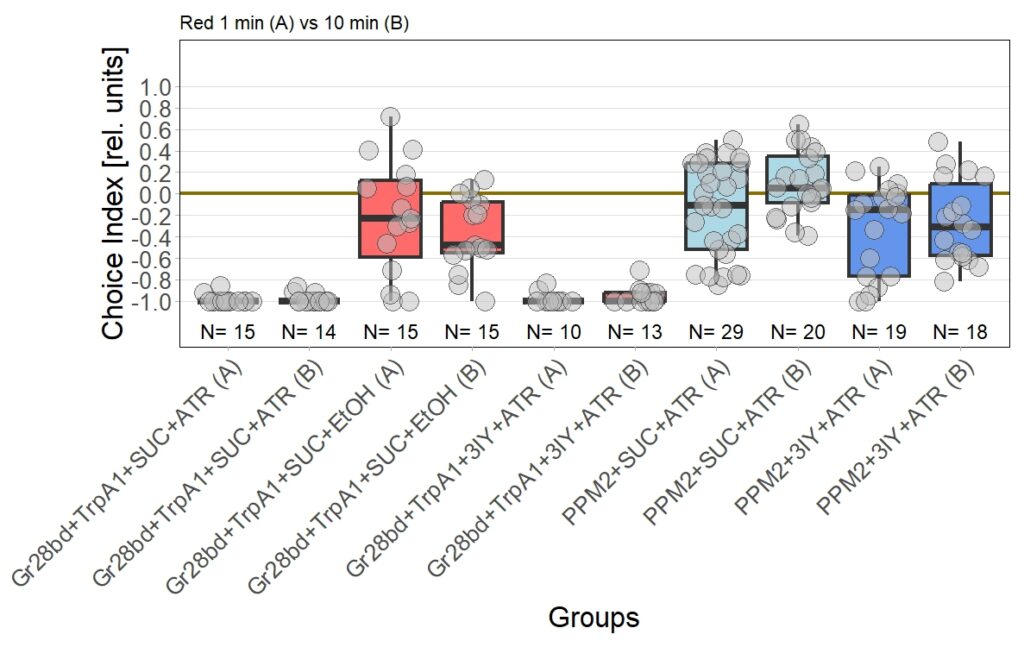
The new set of experiments uses yellow light instead of red light. This experiments are especially interesting, as I observed stronger effects for the experimental group for yellow light.
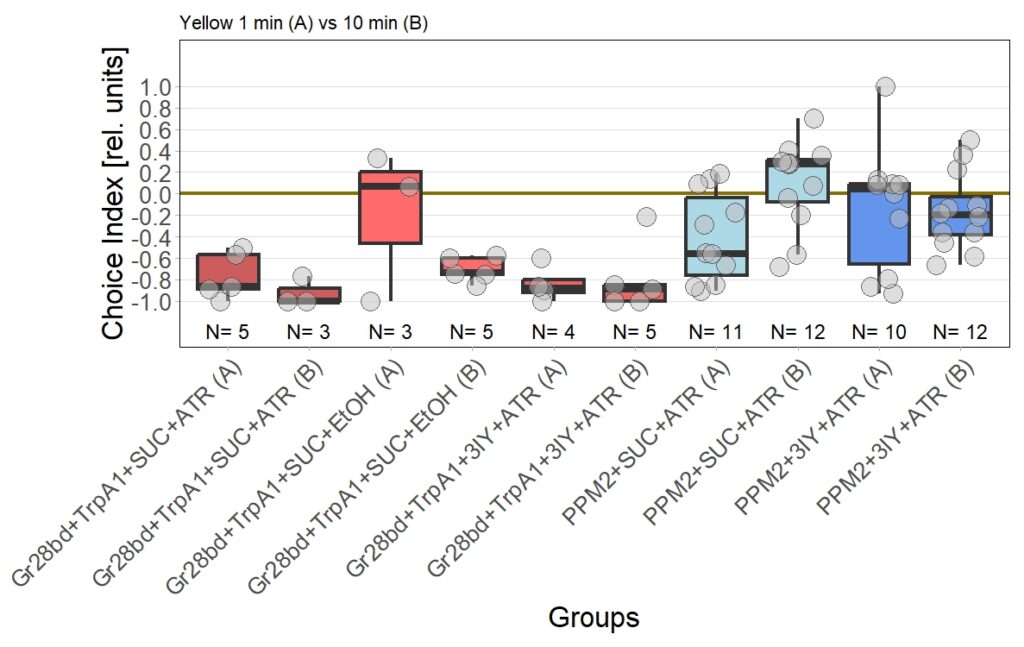
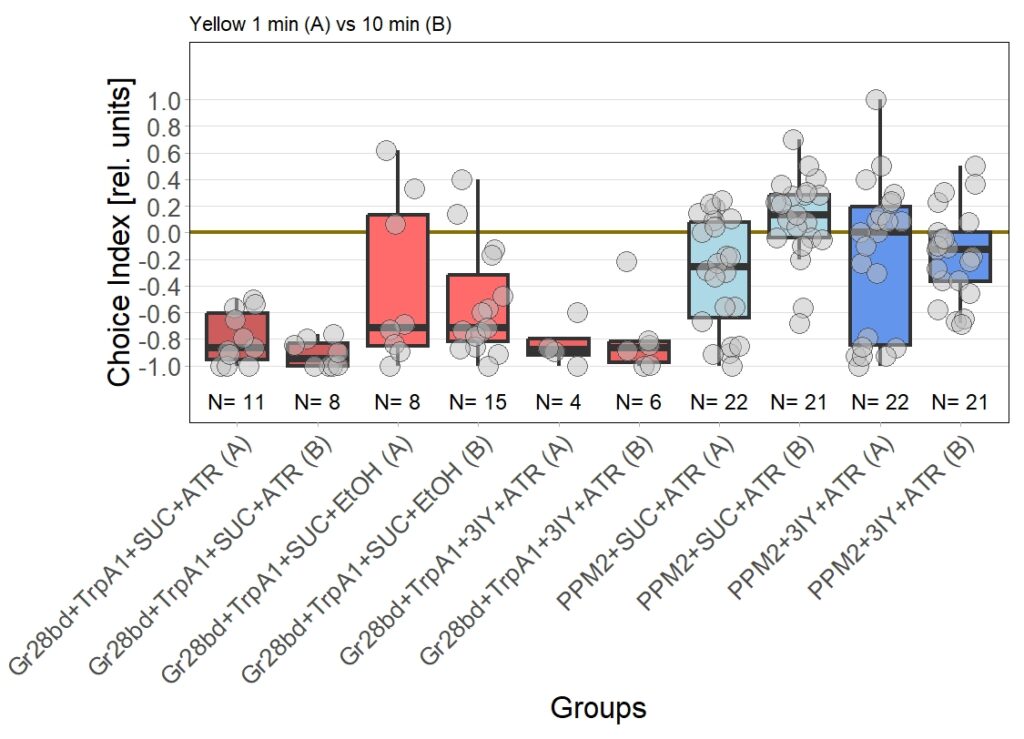
There are two main points to discuss about the data.
First, the negative control (Gr28bd+SUC+EtOH) … I was hoping that I solved the problem with the avoidance in flies that were not treated with ATR. These flies should not avoid optogenetic stimulation since without the chromophore, there should be no, or at least very weak, activation of the CsChrimson channel. Even if the sample size of 5 is rather low, it is a bit worrying that when these flies were tested for 10 minutes ((Gr28bd+SUC+EtOH (B)), they show avoidance comparable to control flies that were treated with ATR and tested for 1 minute ((Gr28bd+SUC/3IY+ATR (A)).
On the other hand, the experimental groups look pretty good. For now, I was not only able to reproduce results from the first 1 vs. 10 minute T-Maze testing with yellow light, it also seems that 3IY-treated PPM2 flies show the same phenotype as when tested in red light.
For the next few weeks I will have to increase sample sizes, aiming for 30 for each of the experimental groups. I will only include a few control groups treated with ATR, as the effect size here seems to allow for a lower N. Presumably, I will include more untreated control flies, to see whether the avoidance will persist or if the current results simply arise from the low sample size.
Update 28/11/25
Category: Biogenic Amines, Optogenetics | No Comments
Updated results from JoyStick experiments with the “new” control driver line (and some OA flies)
on Monday, October 13th, 2025 12:46 | by Daniel Döringer
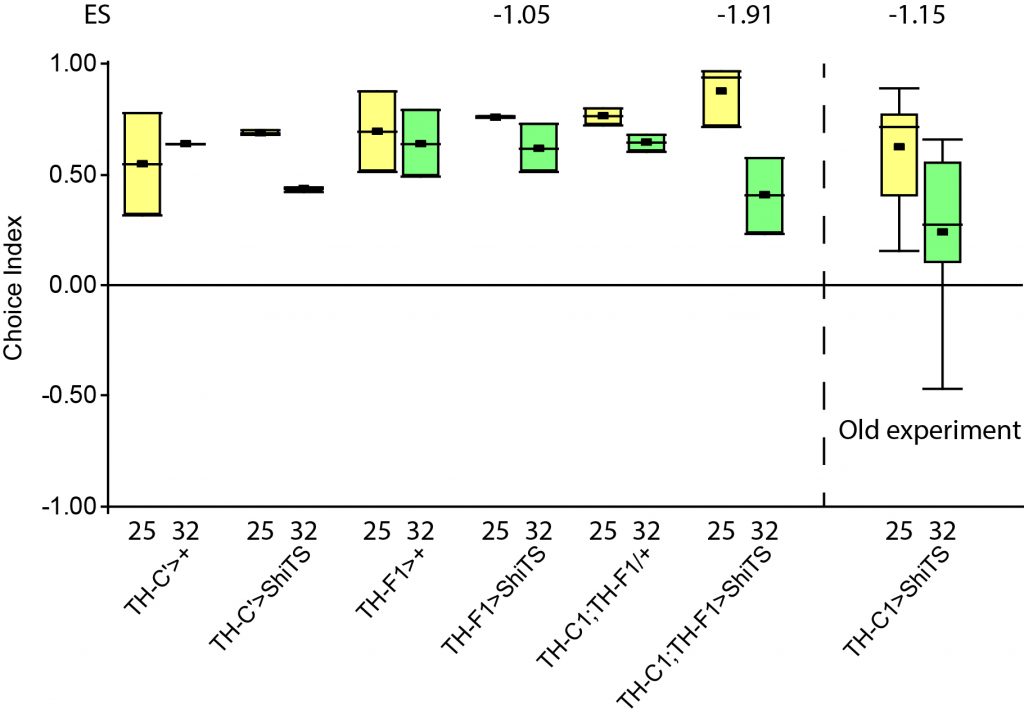
Unfortunately I am still dealing with the problem that flies from my negative control group avoided optogenetic activation, even when their optogenetic channel should not work without ATR supplementation. To tackle this problem I used a “new” effector line (I prepared a new stock from our stock collection) for crossings and tested the offspring, without any improvement concerning the avoidance. Unfortunately the “new” effector line I tested turned out to have lost its “NorpA” mutation which would ensure that the male offspring is blind, thus should rule out any phototaxis-bias. Since flies still avoided the light, a) the effector line does not seem to be the problem and b) the ability to see does not seem to affect the flies behavior in the JoyStick setting.
As a next step I targeted the driver line, as maybe a mutation might have lead to an increase in Gal4. Since we always observed a slightly negative values for our negative control group, hinting that at least some residual activation of the CsChrimson channel is possible even without ATR, an increase in Gal4 might lead to a higher expression of the optogenetic channel.
Residual activation + More channels = More residual activation = Our observed avoidance???
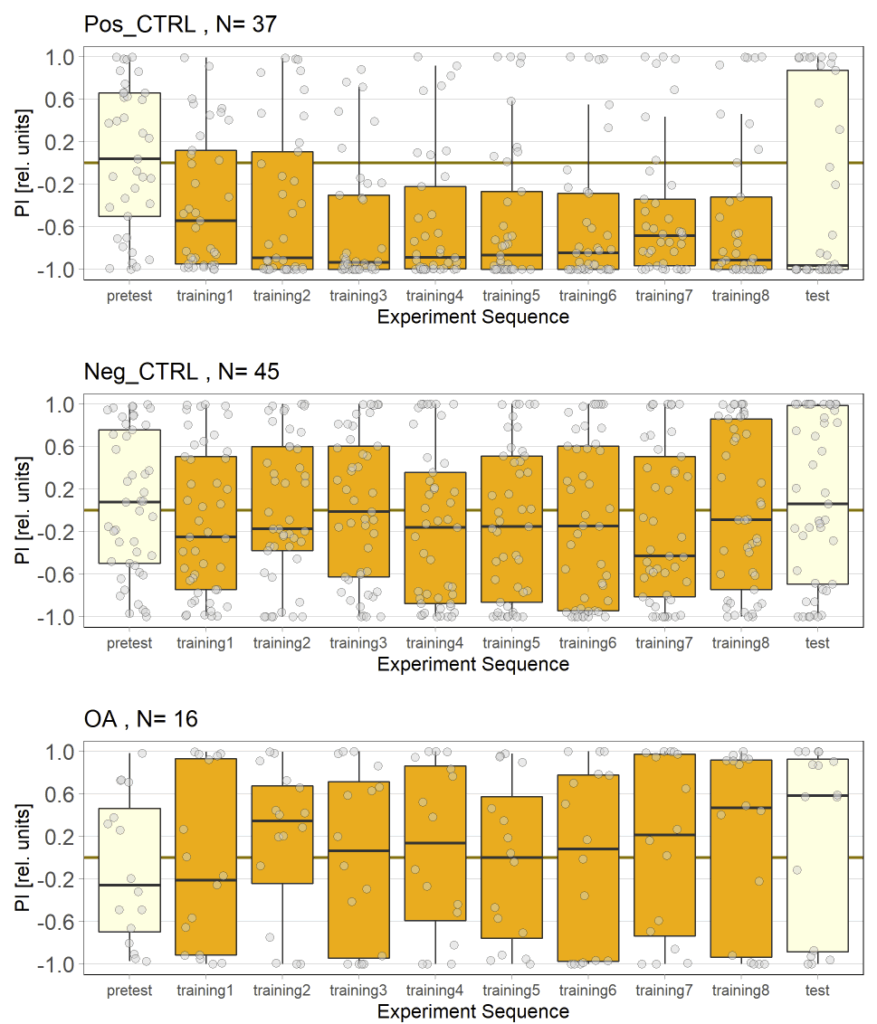
So positive control looks good, negative control is still slightly negative but to an extend that I would consider neglectable.
The additional group here called “OA” are OA;VuMA2 x NorpA, 20xUAS-Chrimson flies. It’s way too early to make any assumptions from the current data but I am looking forward to the results for this group.
Update 27/10/25: Added more flies
Category: Biogenic Amines, Optogenetics | No Comments
Salt (1.5 M) avoidance of 3rd instar larvae
on Sunday, October 12th, 2025 11:53 | by Radostina Lyutova
Category: Biogenic Amines, DAN, Food preference, Larve, neuronal activation, Optogenetics | No Comments
Salt (1.5 M) avoidance of 3rd instar larvae
on Sunday, September 28th, 2025 12:21 | by Radostina Lyutova
Category: Biogenic Amines, DAN, Food preference, Larve, Mushroom Body, neuronal activation, Optogenetics | No Comments
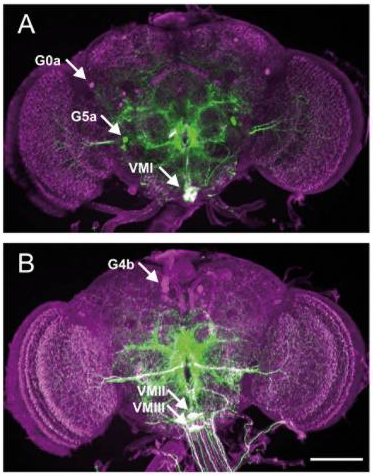
TbH MIMIC
on Monday, October 9th, 2017 2:28 | by Axel Gorostiza
Category: Anatomy, Biogenic Amines | No Comments
Update on DA & photopreference
on Monday, April 24th, 2017 1:55 | by Axel Gorostiza
Category: Biogenic Amines, wing clipping | No Comments
Photopreference shift and DA (new and old GAL4s)
on Monday, April 3rd, 2017 1:56 | by Axel Gorostiza
Category: Biogenic Amines, wing clipping | No Comments
Behavioral Test on DA-GAL4s
on Monday, March 27th, 2017 2:06 | by Axel Gorostiza
Category: Biogenic Amines, wing clipping | No Comments
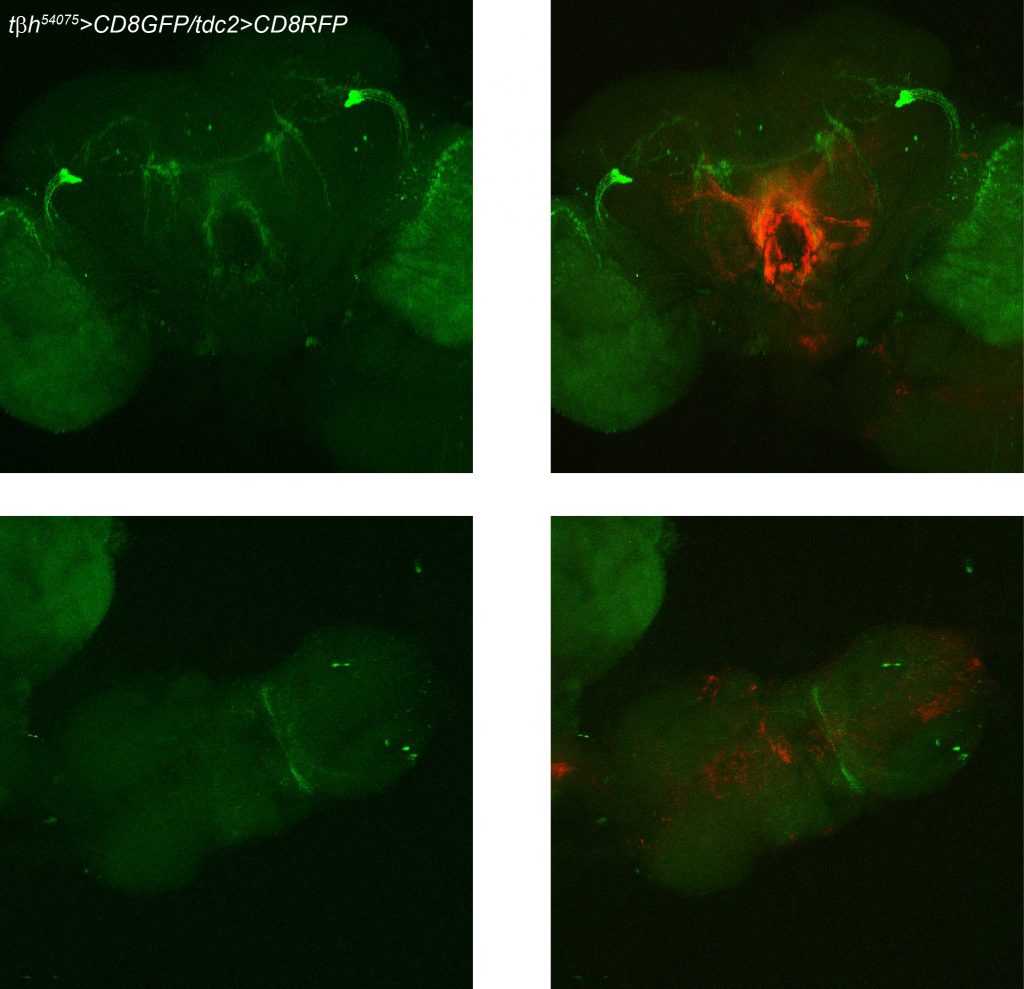
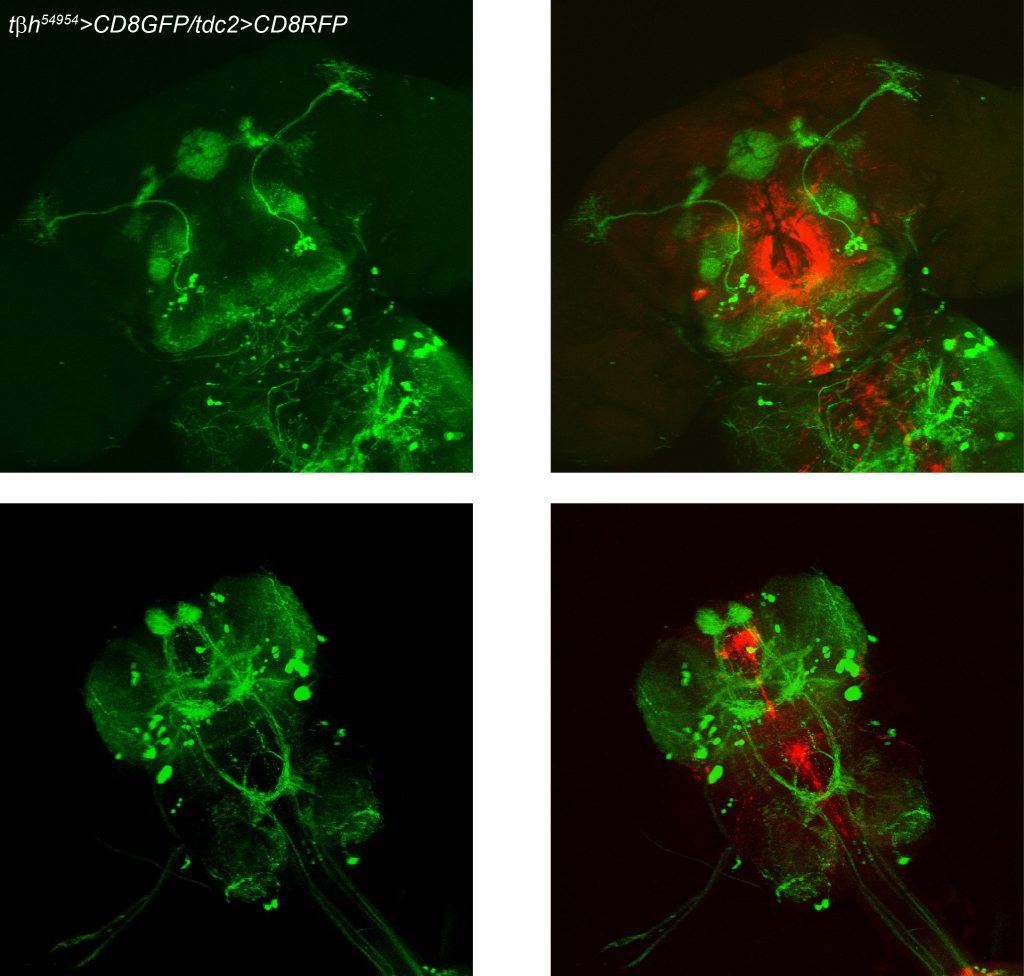
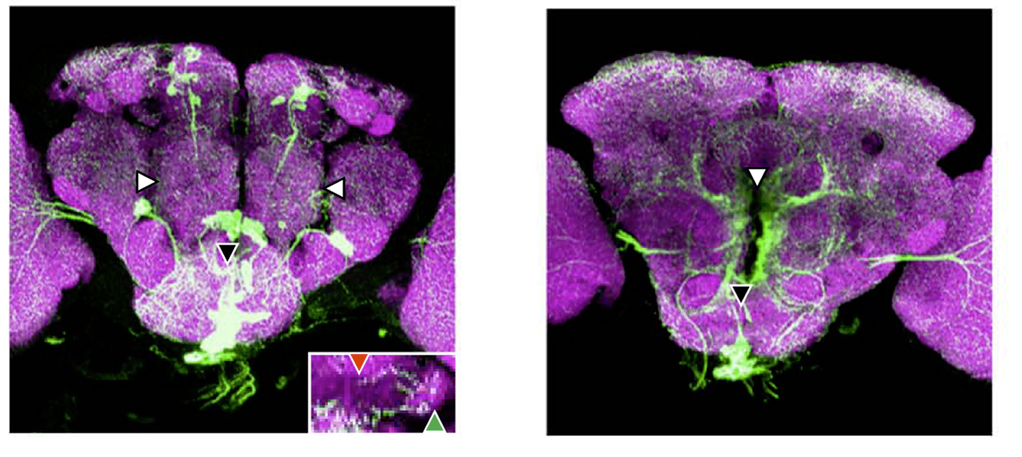
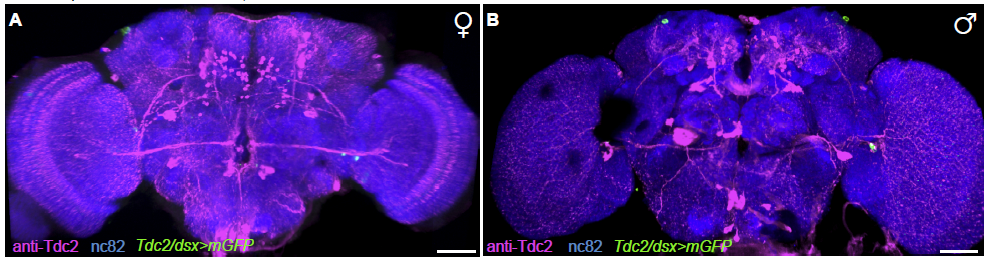
TbH-LexAs and TDC2-Gal4 comparison
on Monday, March 20th, 2017 2:17 | by Axel Gorostiza
TDC2>GFP and anti-TβH (Scholz’s Lab)
TDC2>GFP (Gerber’s Lab)
anti-TDC2 (Goodwin’s Lab)
Category: Anatomy, Biogenic Amines, wing clipping | No Comments
DA neuronal populations and photopreference (counting neurons)
on Monday, March 13th, 2017 3:01 | by Axel Gorostiza
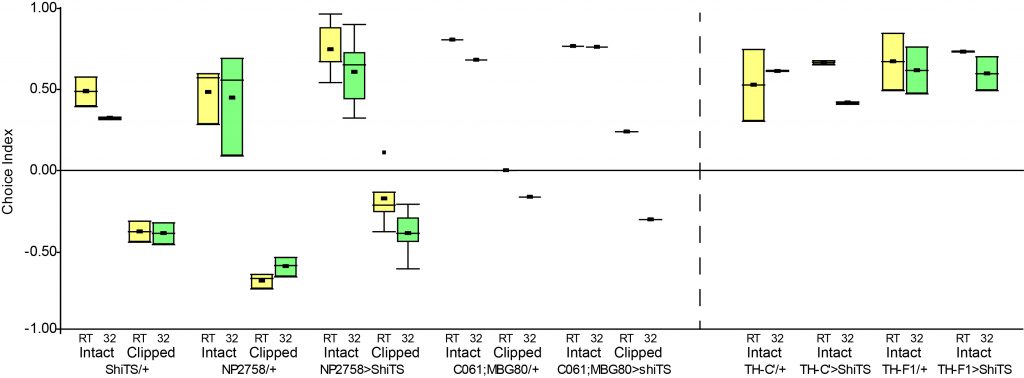
| Genotype | PAM | PAL | PPM1 | PPM2 | PPM3 | PPM4 | PPL1 | PPL2 | VUM |
| thF1>GFP | 0 | 0 | 0,25 | 3,75 | 4,25 | 0 | 3 | 0,75 | 0 |
| thF1;C’>GFP | 0 | 0 | 1 | 7 | 5 | 0 | 2,5 | 5,5 | 1,5 |
Category: Anatomy, Biogenic Amines, wing clipping | No Comments