FENS abstract 2026 Ipek & Friðrik
on Monday, January 26th, 2026 12:58 | by Fridrik Kjartansson
Habit formation circuit in Drosophila Melanogaster
Fridrik Kjartansson*, Ipek Subay*, Radostina Lyutova, Björn Brembs
University of Regensburg, Zoology – Neurogenetics, Regensburg, Germany
*These authors contributed equally.
Classical learning forms association between a novel external stimulus and a
consequence of innate value. Conversely, operant learning associates animals
own behavior to such consequences; eventually habits form which ensure fast
and efficient behaviors. These forms of learning can reciprocally interact with
each other, with classical learning prioritized. A previous study from our group
demonstrated a scenario where in the case of two types of learning paradigms
introduced simultaneously in Drosophila, the classical stimulus was dominant.
However, when the training period was doubled, learning shifted to operant,
allowing habit formation in flies. Moreover, when the prominent associative
learning center, Mushroom Bodies (MB), are genetically silenced, extended
training becomes unnecessary and flies show premature habit formation.
Another study screened Mushroom Body Output Neurons (MBONs), which relay
signals from the MB and found that silencing MBON-02 specifically results in
premature habit formation suggesting MBON-02 acts as final integration center
for the selection between learning types. In this study, we utilize the power of
trans-tango and retro-tango techniques to identify the synaptic partners of
MBON-02 along with validating our results with existing connectomics datasets to
map the whole circuit that regulates the switch between these two types of
associative learning. Through behavioral testing using tethered flight, we will also
narrow down which types of MB cells, called Kenyon cells (KC) , are necessary for
the dominance of classical stimuli in such trials; furthermore, we’ll target
dopaminergic PAM neurons which form feedback loops with MBONs and KC to
elucidate their involvement.
Category: Habit formation, Operant learning, retro-tango, trans-tango | No Comments
Retro-tango confocal images and Torquemeter practice results
on Monday, January 26th, 2026 12:43 | by Ipek Subay
Retro-tango:
(2023) retro-Tango enables versatile retrograde circuit tracing in Drosophila eLife 12:e85041. https://doi.org/10.7554/eLife.85041
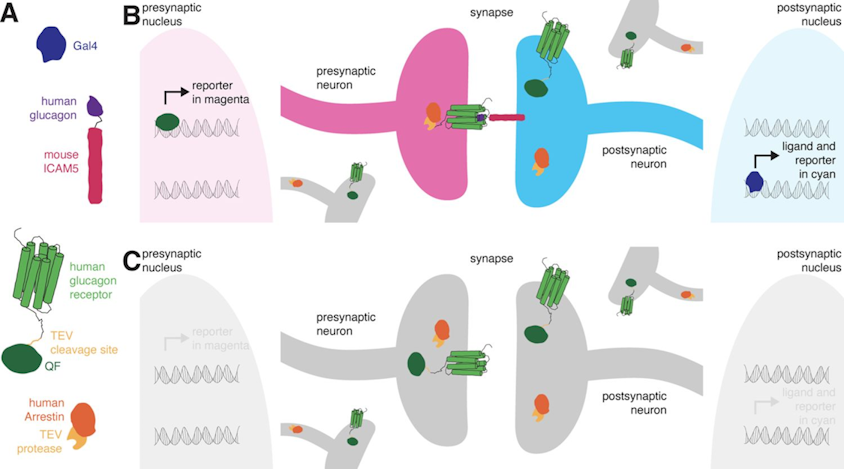
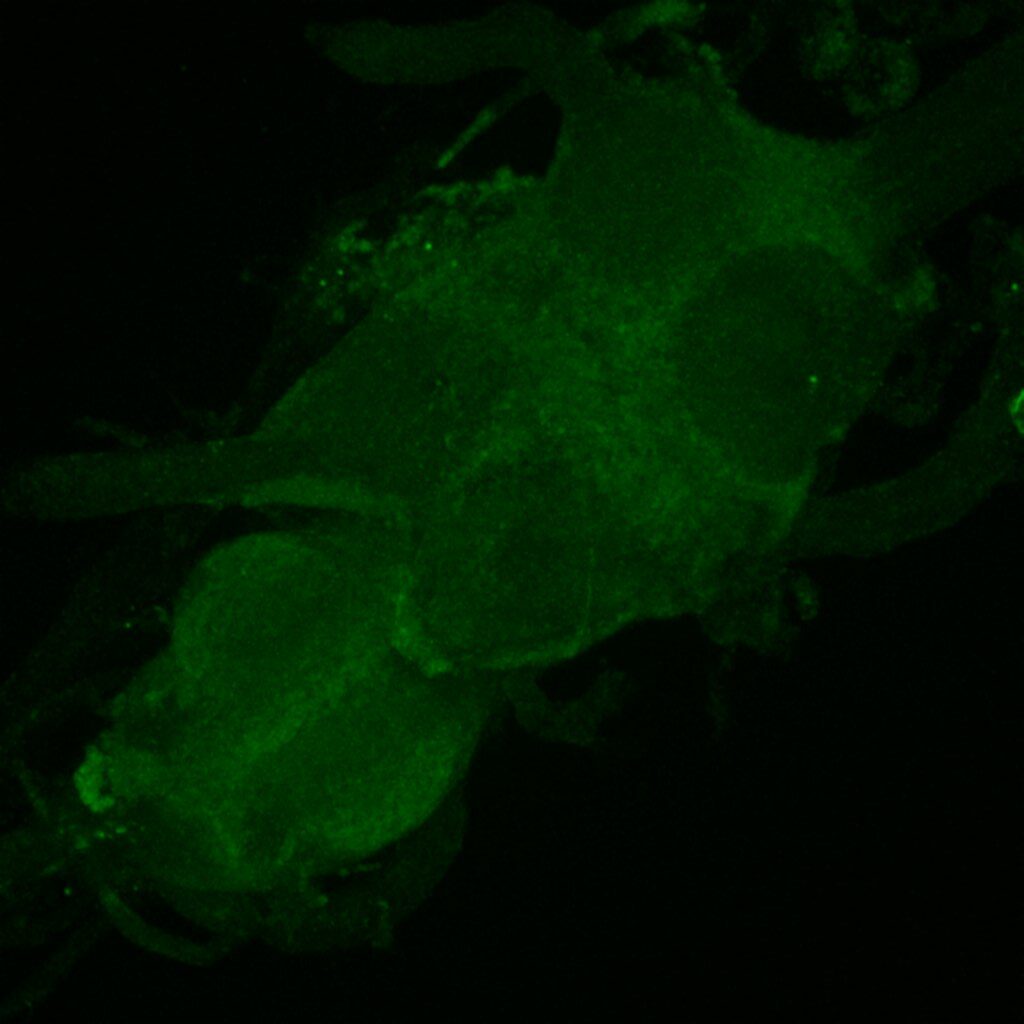
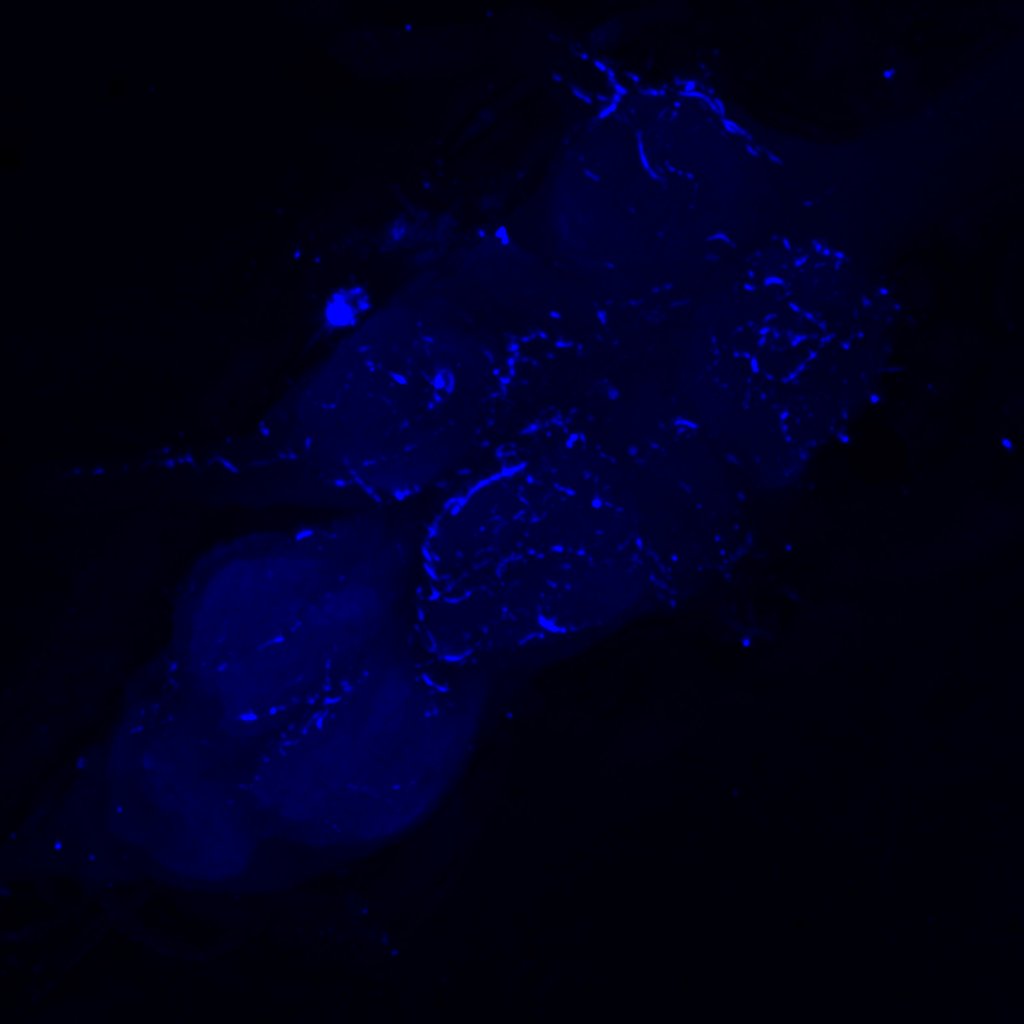
Retro-tango genotype: y[1] w[*] P{y[+t7.7] w[+mC]=QUAS-mtdTomato-3xHA.S}su(Hw)attP8; P{y[+t7.7] w[+mC]=retro-Tango(panneuronal)}attP40/SM6b; P{y[+t7.7] w[+mC]=10xUAS-retro-Tango(ligand)-P2A-EGFP-F}attP2
Crossed with IS69306 (BDSC 601295 and 75552) split-Gal4 driver to express in b1 motor neurons. CNSs stained with anti-GFP Rabbit, (2nd ab: goat anti-rabbit 488), anti-HA Rat (2nd ab: goat anti-rat 555), nc82 (2nd ab: goat anti-mouse 647)
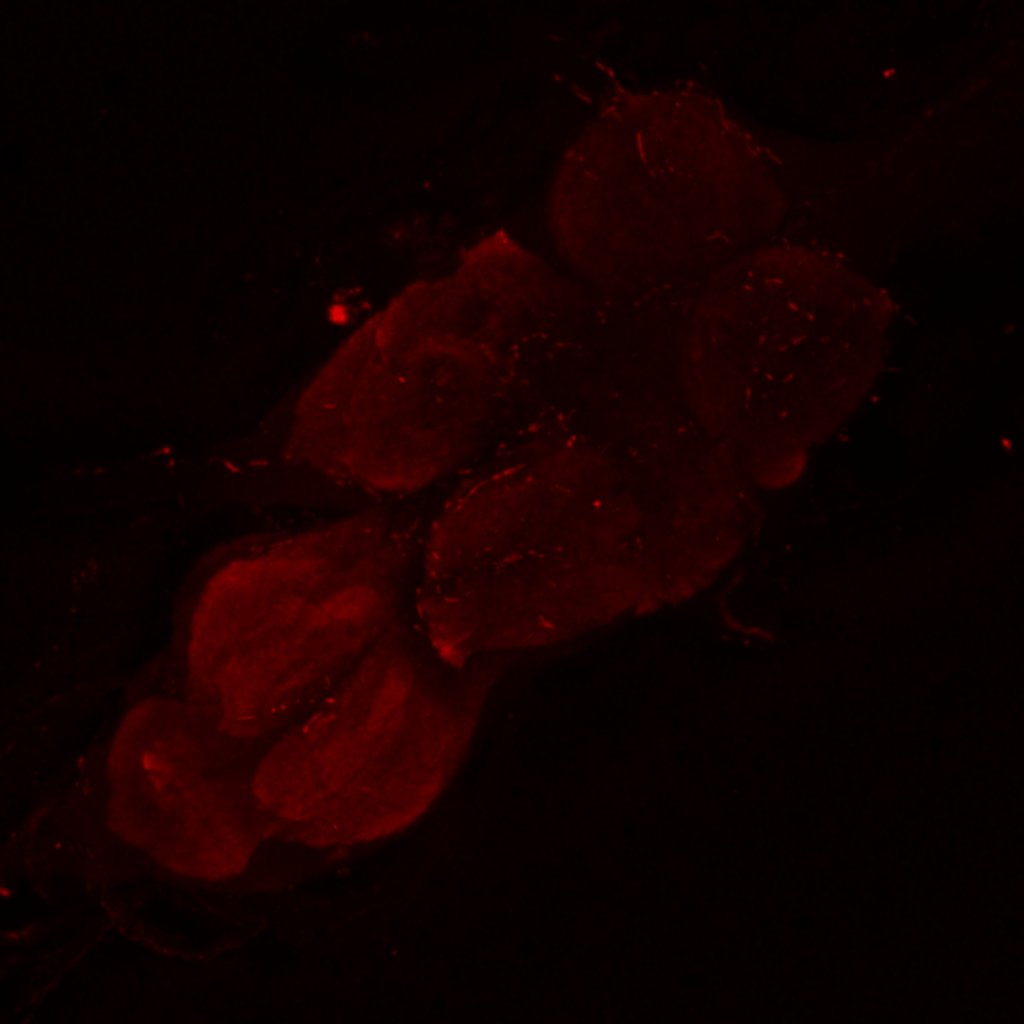
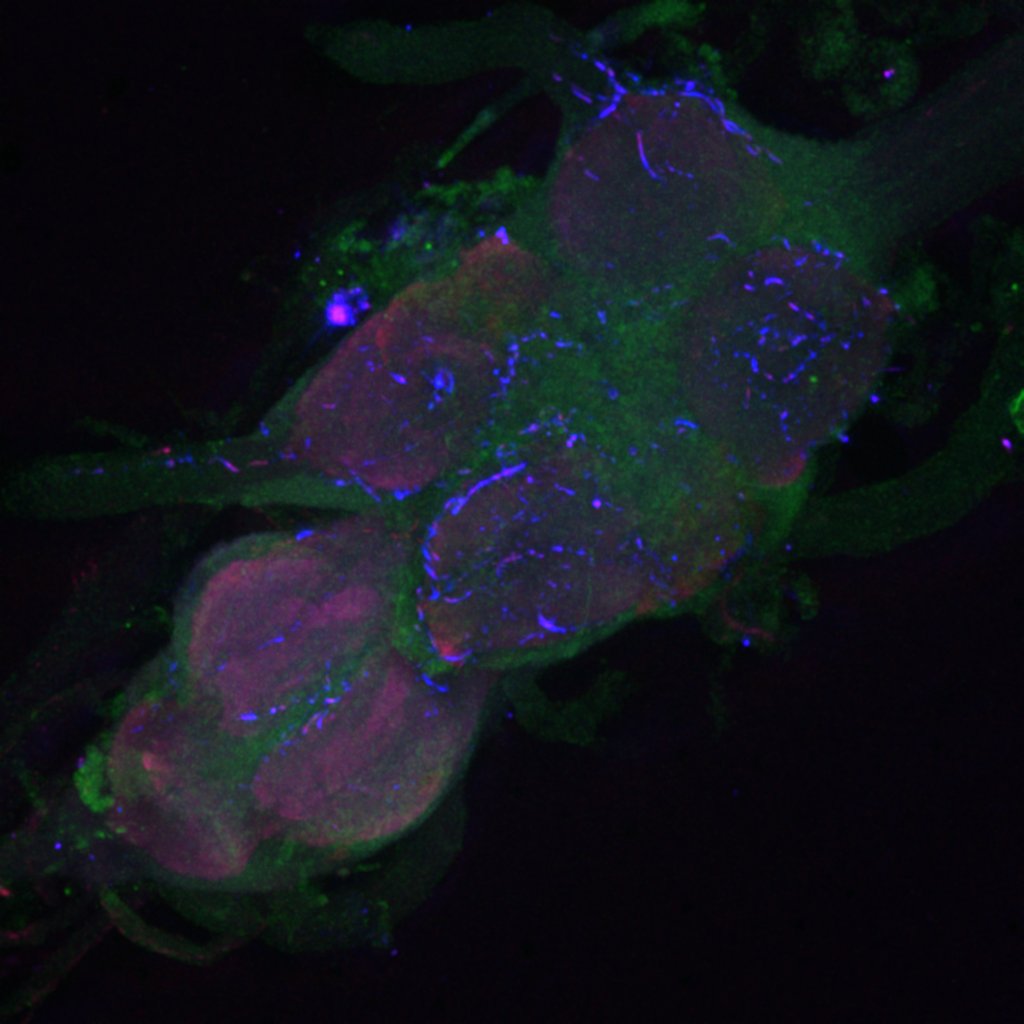
Channel 1 (Green: Laser Line ( 496 nm) Intensity: 23.99%) Spectral Positions/Gain/Offset: (501nm – 556nm) / 865.7 / -0.03
Channel 2 (Blue: Laser Line ( 561 nm) Intensity: 13.10%) Spectral Positions/Gain/Offset: (566nm – 639nm) / 537.7 / 0.03
Channel 3 (Red: Laser Line ( 633 nm) Intensity: 23.99%) Spectral Positions/Gain/Offset: (644nm – 776nm) / 802.9 / -0.01
Second line retro-tango crossed with SS98650 (split-Gal4 driver line from Janelia targeting b3 motor neurons)
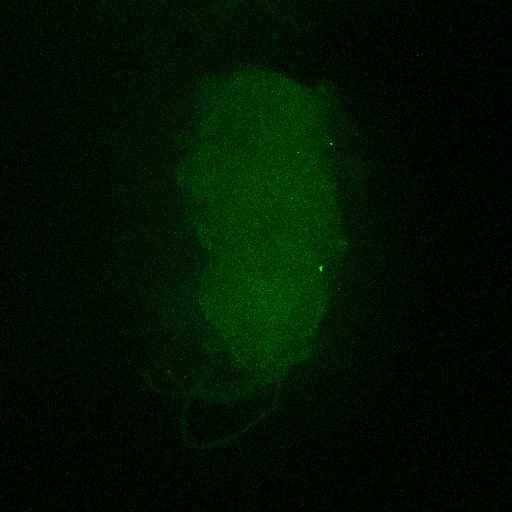
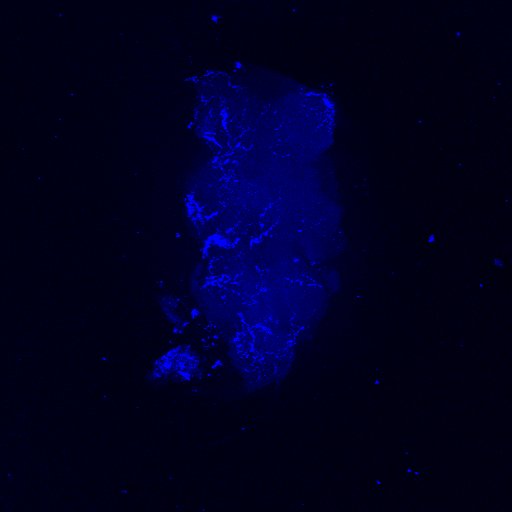
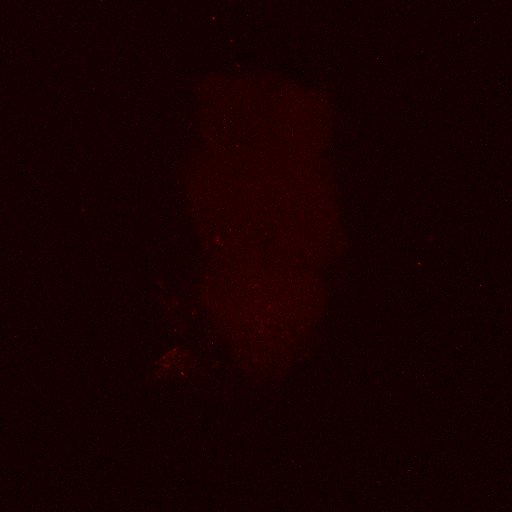
Settings are same as above
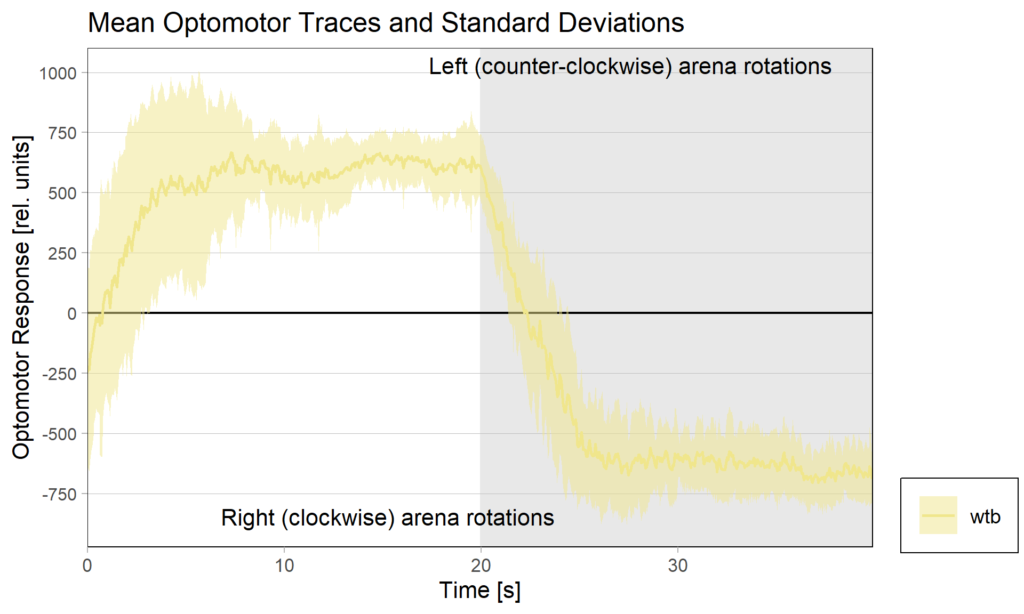
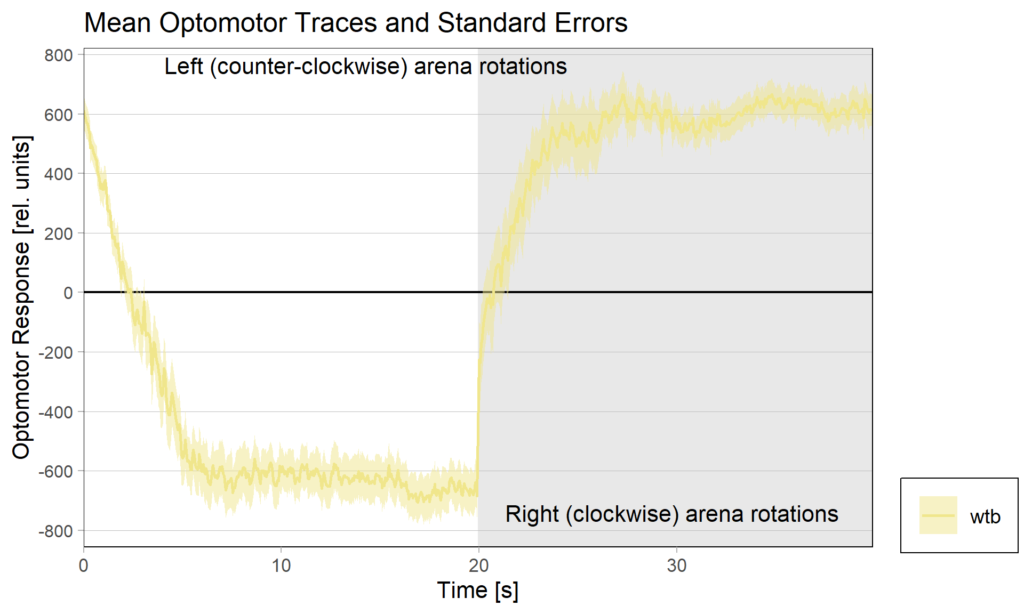
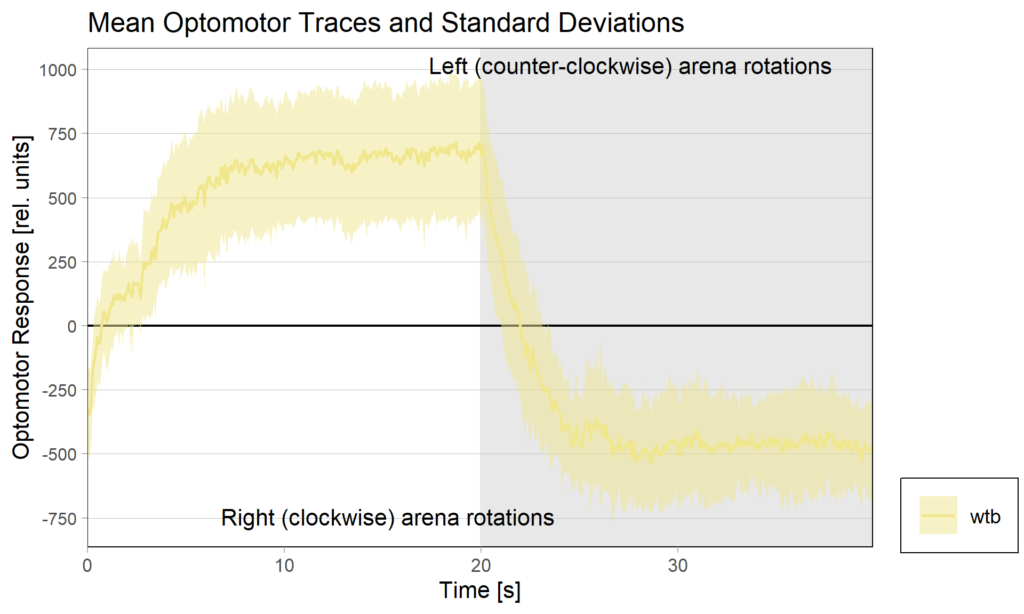
Torquemeter Practice with WTB Flies
N=10 out of 24 glued flies
Optomotor at start:
Optomotor end:
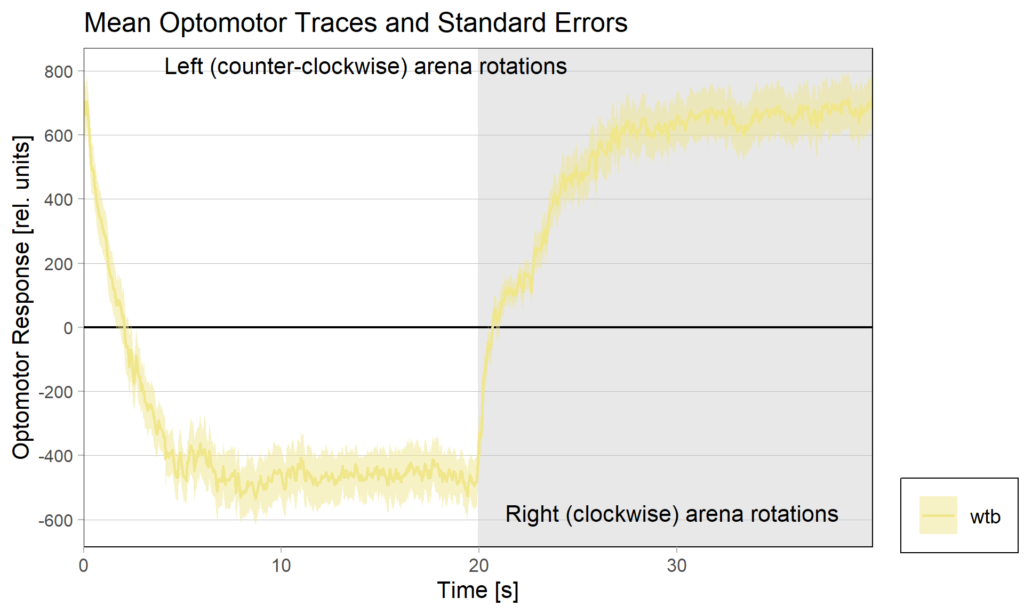
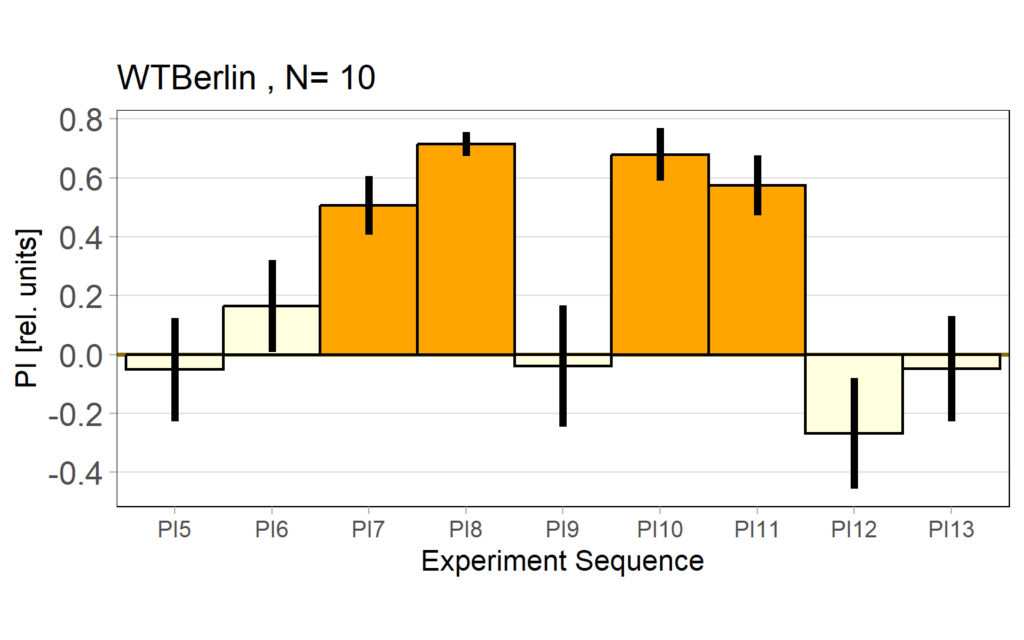
Performance index:
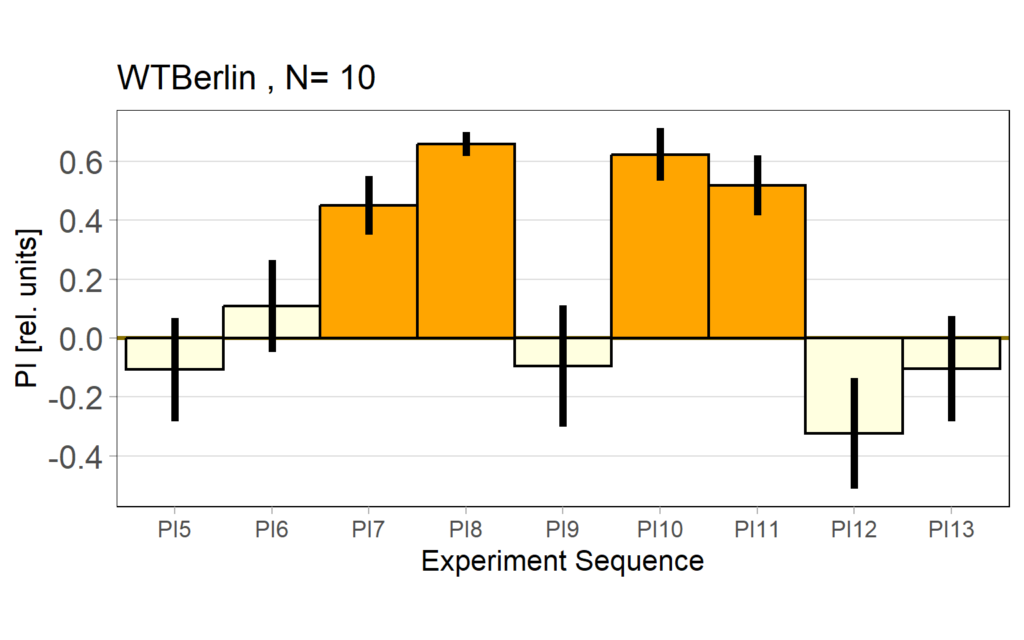
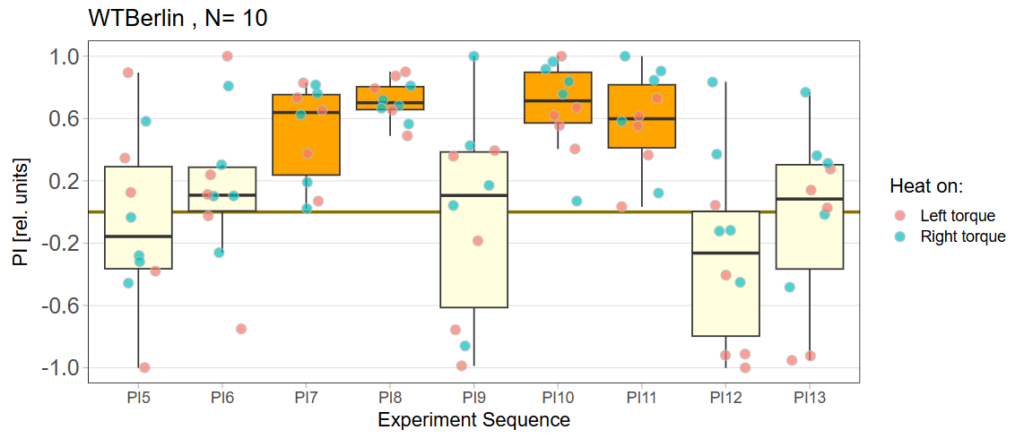
Performance subtracted:
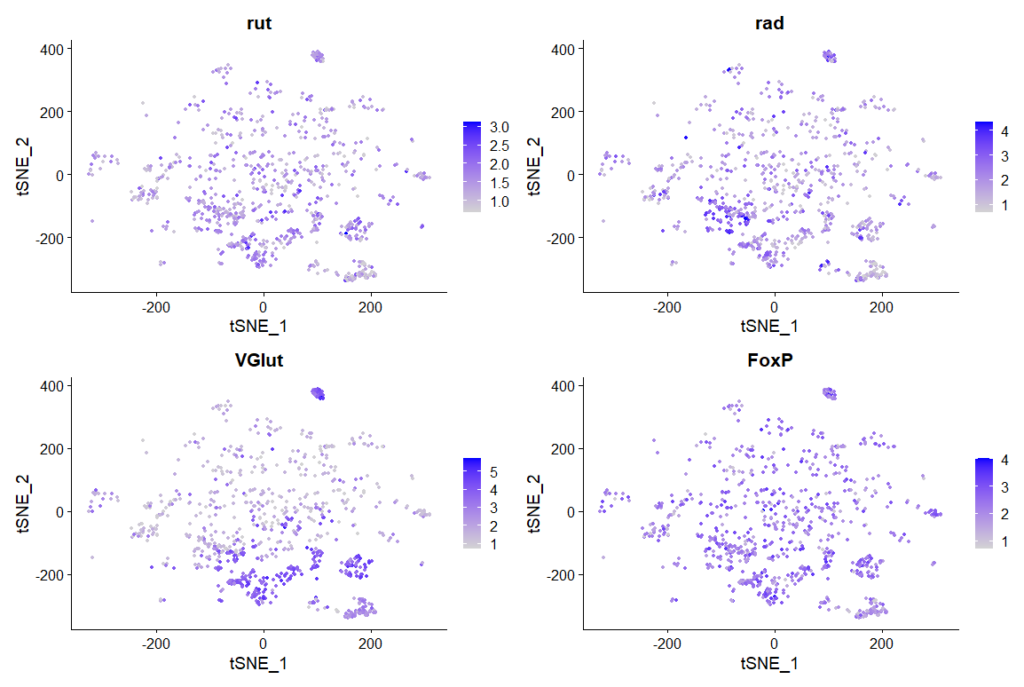
rut and rad expression in ventral nerve cord:
cells that express all 4 genes more than 2 fold, 889 cells in total
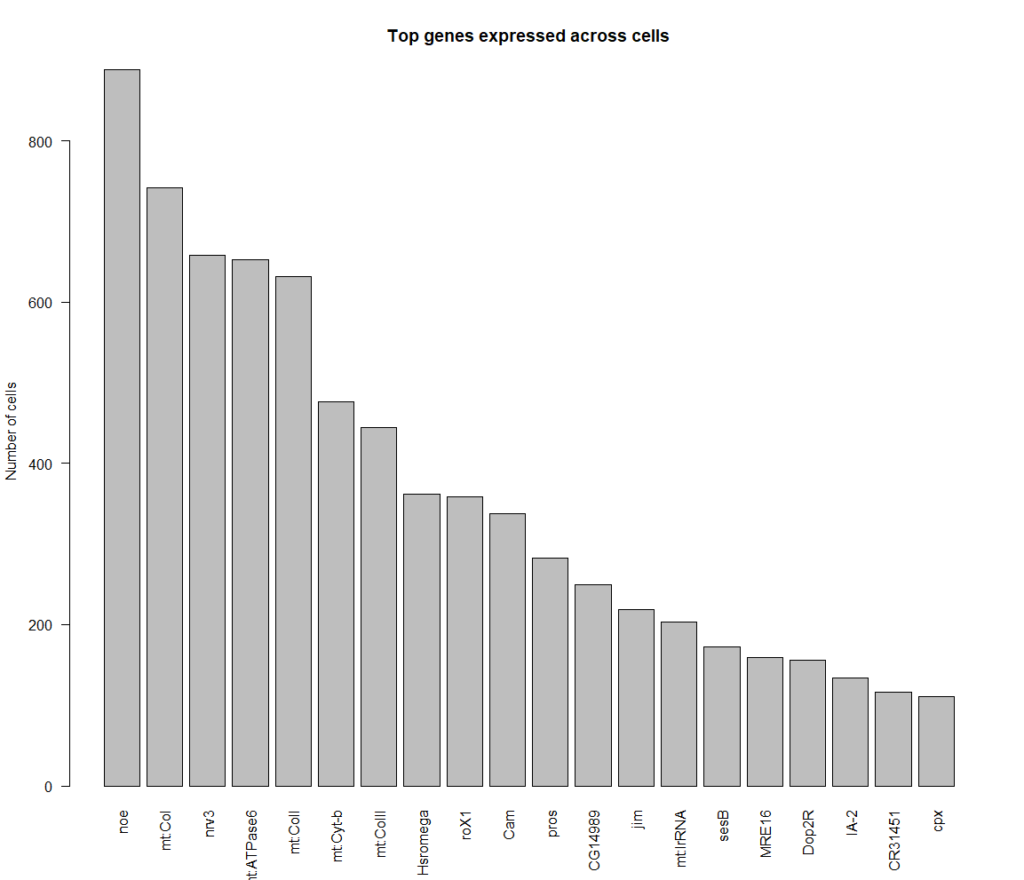
Top genes expressed in all of these cells:
Category: Habit formation, retro-tango | No Comments
