Construct for FoxP isoform B ready
UNIVERSITY PART
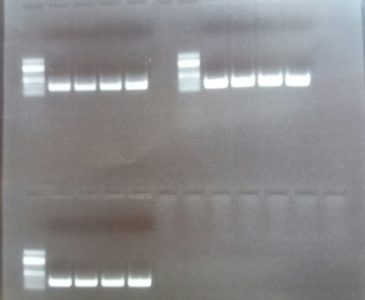
The image shows the presence of the HomI insert (upper bands) in a vector where we have already ligated the HomII insert for the FoxP isoform B. This is the construct that will allow us to insert the Gal4 in the ORF. This construct is thus ready to be injected
Construct for FoxP conditional KO
UNIVERSITY PART
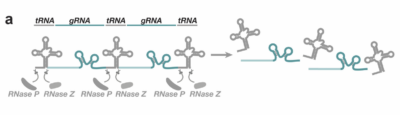
How the technique works: tRNA based vectors for producing multiple CRISPR sgRNA from a single transcript. Using multiple sgRNA will augment gene repression. In this technique the endogenous tRNA processing machinery liberates multiple functional sgRNA from a single precursor transcript in the nucleus. The vector used (pCFD6) permits cloning of several (three in this case) sgRNA flanked by Drosophila tRNA downstream of a single promoter.
The conditional KO can be done by using UAS-tRNA-sgRNA constructs.
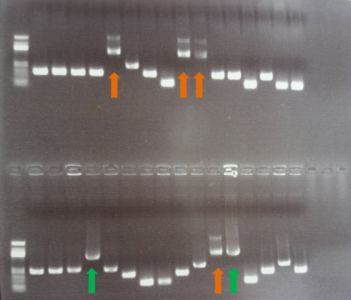
PCR amplification product of three different FoxP fragments, those fragment will be ligated in only one appropriate vector.
PCR of the tentative of ligation of all the 3 fragments above: green arrows indicate clones that should be having incorporated all 3 fragment, orange arrows indicate clones that can be positive to all of 3 fragments but have a strange double band (the sequencing step will tell us the results).
KLINIKUM PART
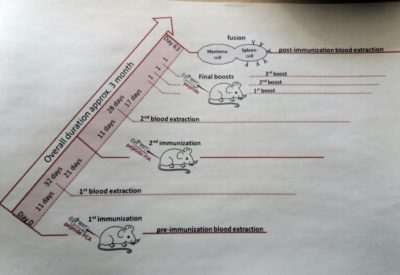
Protocol for mice immunization for creating monoclonal Ab
New cloning plasmid for FoxP homology domains
UNIVERSITY PART
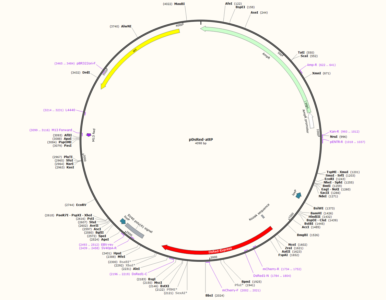
Map of the phd-ds-Red-attP plasmid used for cloning: 1 desired construct is aiming to target all dFoxP isoforms with one homolgy domain starting at exon 1 to exon 3, the second homology domain starting at exon 3 to exon 6. The second construct is aimed to target just dFoxP IsoB, with both the homology domain at the end of the protein. This plasmid is aimed to knock down the FoxP gene without the Gal-4 knock-in.
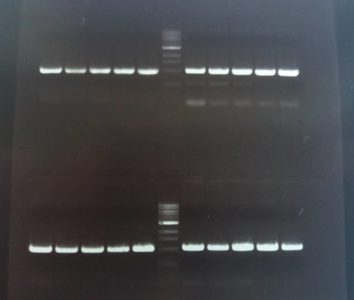
PCR product of the homology domains 1 and 2 for FoxP (upper part of the image, left and right), and homology domains 1 and 2 for FoxP-isoformB (lower part of the picture, left and right): amplification of the desired homology domains via PCR (5 replicates for each construct in order to test different annealing temperatures). This products are going to be used for the subsequent cloning in the plasmid.
Starting of the FoxP project
UNIVERSITY PART
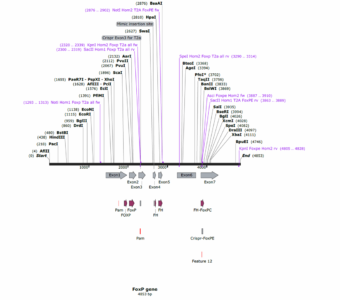
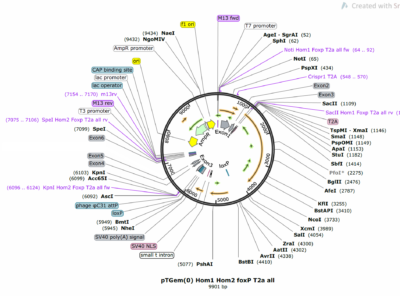
Maps of the FoxP gene and of the pTGem plasmid used for cloning: 1 desired construct is aiming to target all dFoxP isoforms with one homolgy domain starting at exon 1 to exon 3, the second homology domain starting at exon 3 to exon 6. The second construct is aimed to target just dFoxP IsoB, with both the homology domain at the end of the protein.
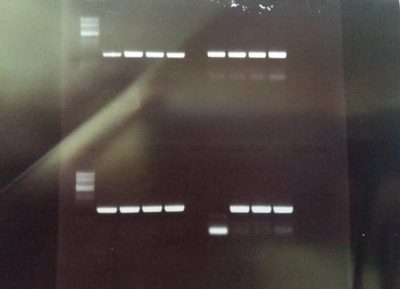
PCR product of the homology domains 1 and 2 for FoxP (upper part of the picture, left and right), and homology domains 1 and 2 for FoxP-isoformB (lower part of the picture, left and right): amplification of the desired homology domains via PCR (4 replicates for each construct in order to test different annealing temperatures). The products will be used for subsequent cloning in the vector.
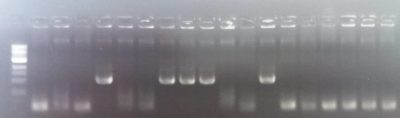
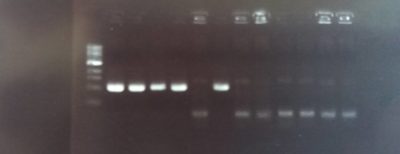
Plasmid with Hom1 Hom2 FoxP ready: the image shows the presence of the homology domain 2 for dFoxP in a plasmid where we already ligated the homolgy domani 1. This construct is thus ready to be injected.
KLINIKUM PART
We transfected E.coli cells with the plasmids for IsoA and B from the Schaff lab (Berlin) and subsequently we sequenced them with 4 primers each (1 FW primer sequence in the plasmid upstream the ORF and 1 RV, 1 FW primer sequence in the middle of the ORF and 1 RV).
pcDNA plasmid IsoA correctly amplified in E.coli
pcDNA plasmid IsoB correctly amplified in E.coli