August
on Thursday, August 27th, 2020 1:46 | by Ottavia Palazzo
- Halfway in analyzing the Buridan data altogether (not in two batches)
- Maxiprepped plasmid for making FoxP protein at the klinikum
- Writing the thesis a little bit
- bought Fas-II antibody
________________________________________________________
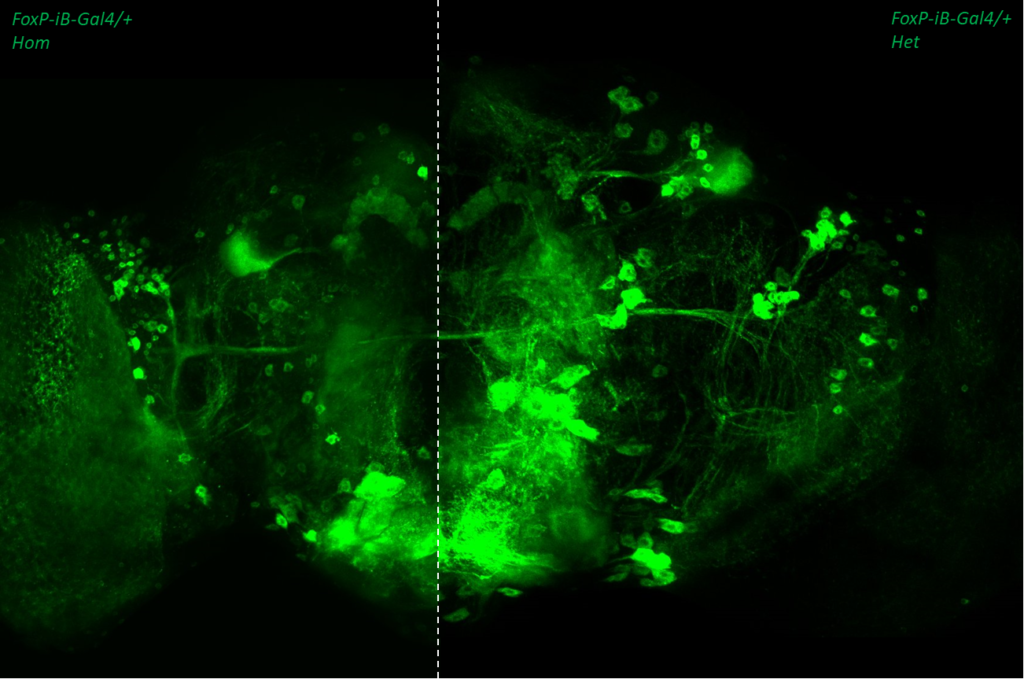
- data 1): FoxP-iB Heterozygous/Homozygous comparison with Stinger-GFP. This time i was cautious with everything: fly all the same age and sex, same larvae density, same number of copies of GFP
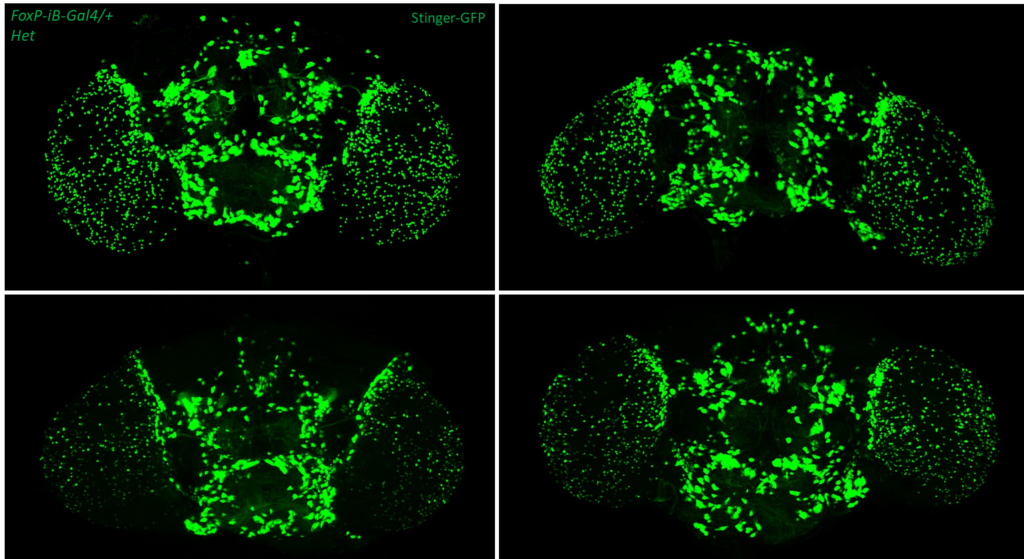
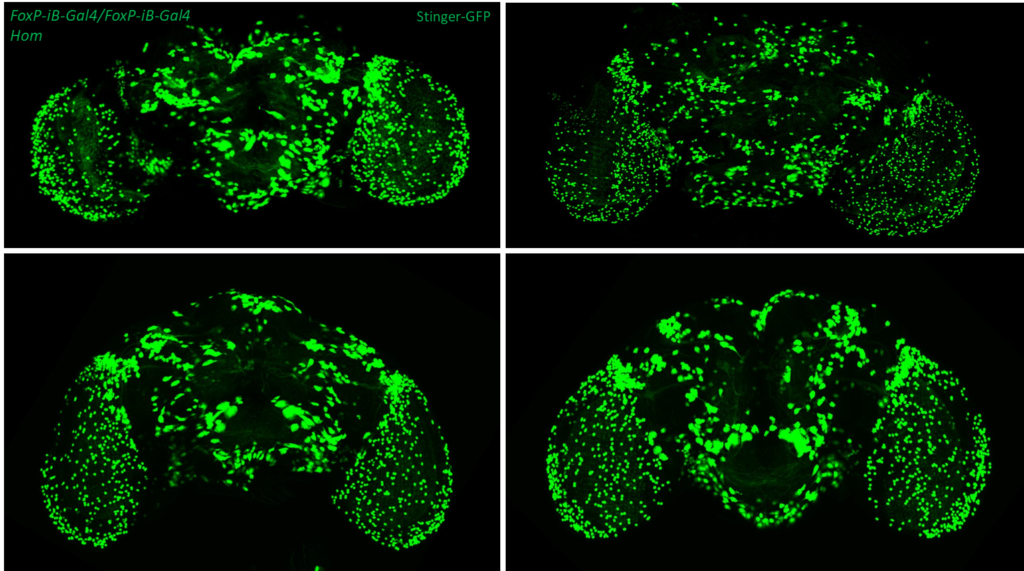
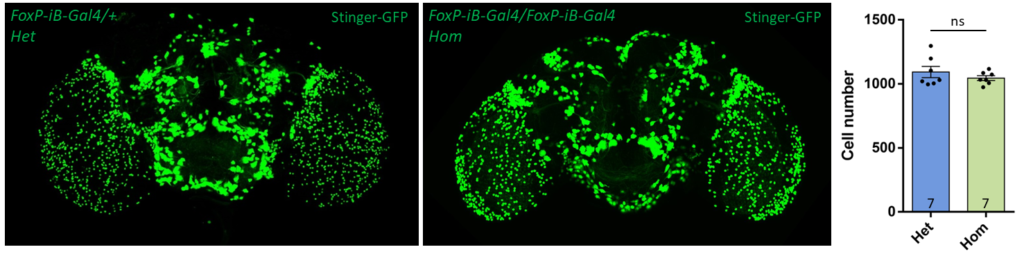
I can not detect any difference between homozygous and heterozygous mutants. I also counted cells in IMARIS and I detected no difference in number or distribution.
- data 2): FoxP-iB Heterozygous/Homozygous comparison with CD8-GFP. This time i was cautious with everything: fly all the same age and sex, same larvae density, same number of copies of GFP
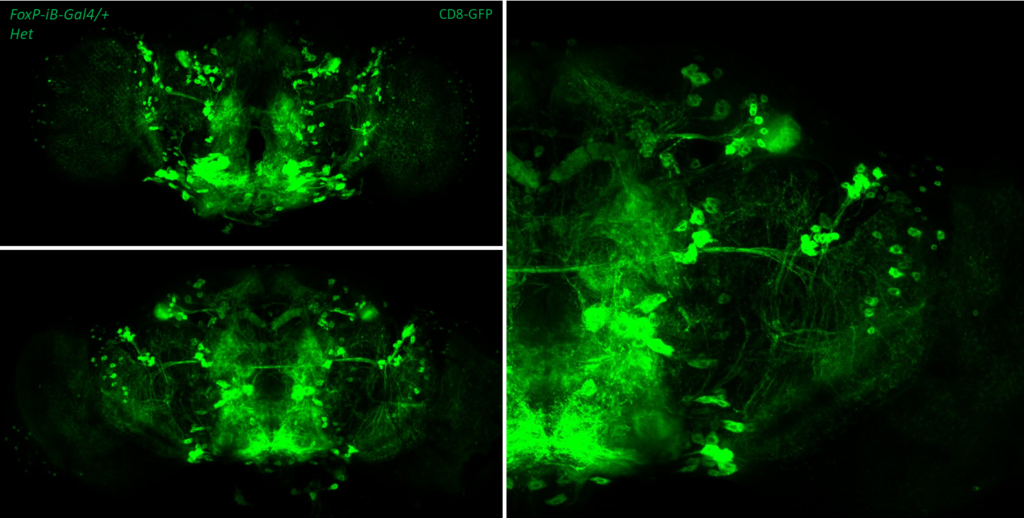
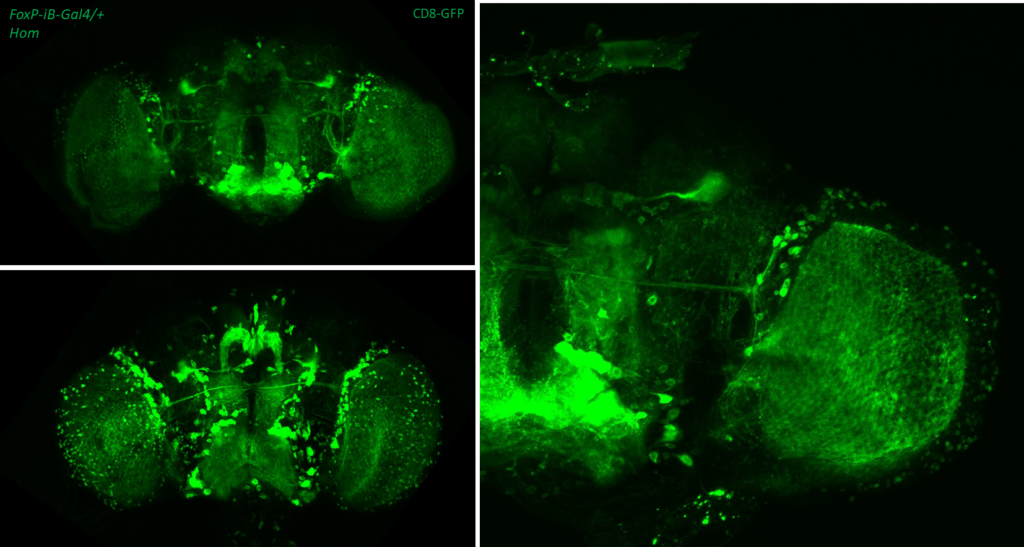
I can see some differences but i do not know how to quantify/explain them. Also I am not sure if it is a problem of dissection/mounting. I am not convinced.
- data 3): nc82 staining on homo/hetero FoxP-iB. I do not see differences

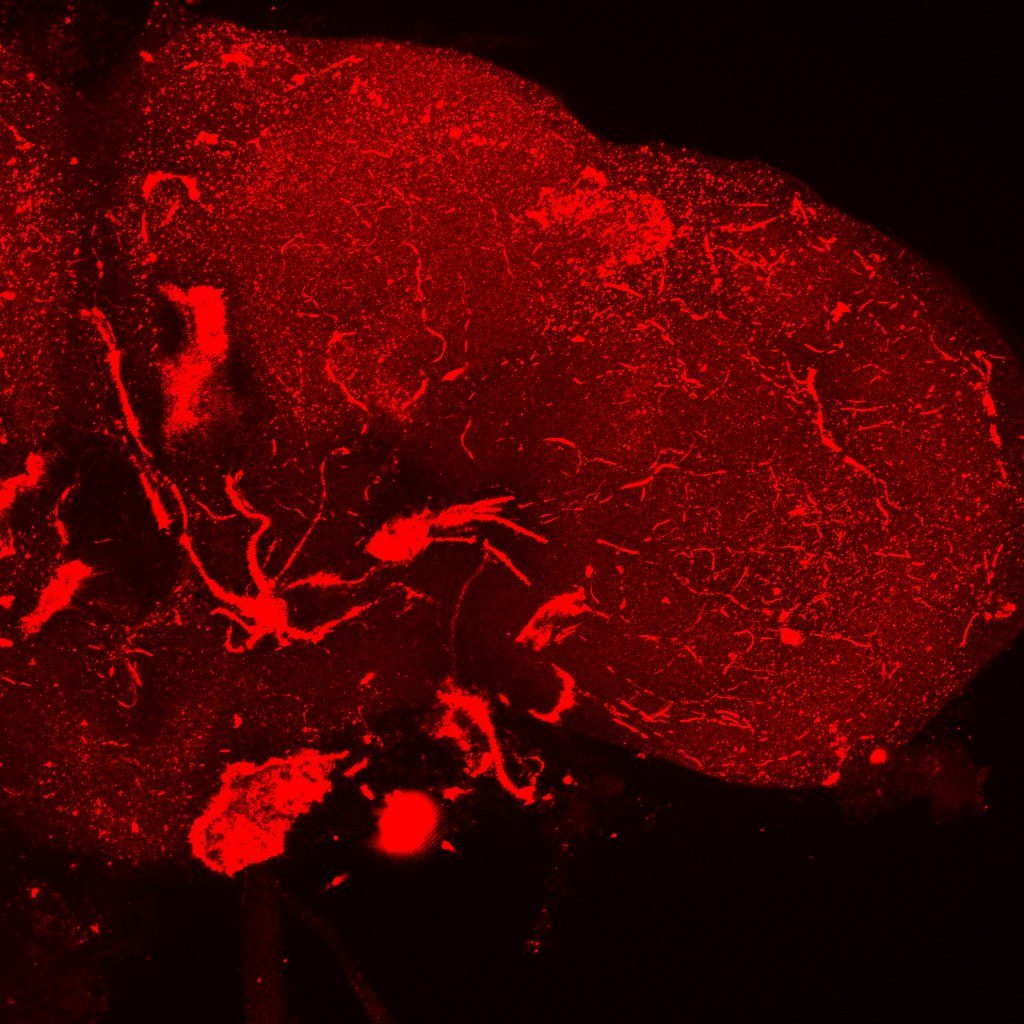
Problem: I haven’t managed yet to make the FasII antibody work:
I have used a 1:10 concentration of primary antibody for one night and a 1:100 concentration of secondary antibody for 4 hours. I will ask someone
In the picture I have increased the gain a lot just to be sure. I could not see anything
Category: Foxp

Leave a Reply