UAS-PKC53eRNAI
So I got new flies to test. After one day of testing, I had only one control fly going through the test, all the 5 other were dead. That one did not learn.
I thus confirm that there is a problem with these flies. This means that I have no phenotype with one RNAi, and one phenotype with the other non-triggered RNAi which is probably not self-learning specific. This also means that the RNAi induction may not be sufficient and that I have no positive control for that.
The RNAi data are therefore not conclusive at all (neither positive nor negative results can be linked to any gene).
PS: I have checked the flies and overexpression of GFP did occur after 2 days Heatshock.
I will check the mutant again next week and I will be over with learning experiments. Still need some anatomy but then, I can finish the paper.
not enough data to be sure- new cross needed
I will first test the mutant, if there is a phenotype, I will cross the RNAi lines to d42Gal4 to avoid using temperature shifts, which seems to be deteriorating the UAS RNAi vienna control line (only 4 flies made it to the test, lots of them died during the overnight heat shock).
looks good, hope to confirm that in the following two weeks
https://figshare.com/preview/_preview/658786
It would be great to have some physiolgy data (electrophy) to have a guess about the effect of the PKC in the motorneurons (?)… Do you know if there is a paradigm to look at LTP and LTD at the MNJ?
publishing torque meters comparison
here is the draft: https://figshare.com/preview/_preview/644625
Is there any good publication describing the wing beat analyser?
other remarks?
Science and semantic web
I was on a meetup of corporate semantic web last Tuesday. These people are using semantic web technologies (making machine readable content based on ontological terms and relation between these terms) to improve the efficacy of private companies. For instance, they work on ways to improve wiki contents which may be produced in a company. This corresponds at using ontological term to annotate the wiki content and other related technologies. This can be used to find an expert in one category (=somebody who’s posts are rarely corrected on a specific subject).
What is the scientific community (the one which should be leading the way actually) doing during that time: we use text search in “keywords” and titles to find the appropriate literature, that we have to read thouroughly to drive our one conclusions about these different parts… At least, that is what we do 90% of the time, and we all know how inaccurate this can be. Experimental results may be translated into a machine readable content, why aren’t we doing it (it could make everything that much simpler, faster and more accurate)?
The answer: 1. there is no tool nor database where we could do it. 2. Scientists do not have the time to do it, they are over-pressurized to produce data, not to make it reusable or machine readable.
How to push people to use the semantic web technologies, how to ease this use, should it be done by the authors or by the community, pre or post publication, what ontology tool to use,… What can we do? Is anyone asking these questions around? Does a platform like researchgate be a way to introduce this, or should we go for a public solution, inside pubmed for example?
Is any of you asking/answering these questions?
By the way, this post is tagged by none-ontological terms, a shame?
PKC53e mutant
https://figshare.com/preview/_preview/106910
reminder:
username colombj@zedat.fu-berlin.de
password: neurobio
definitive answer should come end of next week.
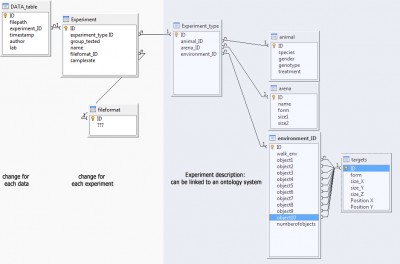
Trajectory data: database structure
CeTrAn is our software to analyse trajectory data, written in R it is free and open source . It was designed to analyse data obtained in the Buridan’s experiment setup. I am now trying to have a larger scope and incorporate different type of data:, for instance:
– Buridan’s experiment done with a different tracker
– Walking honeybee tracking in a rectangular arena, with a rewarded target
– Animal (flies/bees) walking on a ball, using open- or closed-loop experiment setup
– trajectory data obtained from the pysolo software (flies)
– larval crawling data
I want to include an automatic depository of the data in a database. Automatic entries in Figshare is for instance possible. (see older posts). My problem is to find a way to treat the data such that:
1. the raw data is uploaded
2. all data is uploaded also if we use only the centroid displacement (in some data file the head position is also given)
3. the data can be reused and data obtained in different lab, animal, setup can be compared. (data should be organized such that it can be searched and queried).
4. probably other elements that I do not think of….
My main problem: I have nearly no experience in data management/design, ontology or semantic web. Here is a first draft of a database structure that I have thought of. Every feedback would be welcome:
c380 driven PKC inhibition
Unfortunately, the amount of data is a bit too low to draw conclusion, but if one look at the data for each fly:
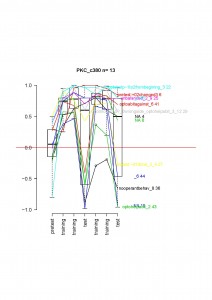
Only one fly not learning and the median would be 0. It looks very similar in the d42xPKCi cross, suggesting that the flies will also show the phenotype.
This will need to be confirmed next year with a new cross.
Other data visible at https://figshare.com/preview/_preview/104549
and later at doi: 10.6084/m9.figshare.104549
following comments
I have just realized that there is no “new comment” section, so we have access only to the last 3 comments. Since there is also no way to have them as rss feeds (only public posts are feeded), it makes it possible to loose track of comments, if somebody (like I just did) look and comment the posts only once in a while.
Is there a way to manage this?
Crosses to check the TARGET flies
Since we have problems with Madeleine’s project, I will make crosses to test that the flies are ok before using them for PKC RNAi experiments.
the putative elavGal$;tubGal80ts flies will be crossed to w; AKAP/Cyo flies. Then the son ElavGal4/Y, tubGal80ts/+ will be crossed to w.
Only non Cyo male flies should get white eyes, if not, the stock is not clean.
In parallel, I will also start crossing nsybGal4 into the tubGal80ts line (2 to 4 crosses necessary depending on the eye color of the heterozygotes).