Trajectory data: database structure
on Thursday, December 20th, 2012 5:16 | by Julien Colomb
CeTrAn is our software to analyse trajectory data, written in R it is free and open source . It was designed to analyse data obtained in the Buridan’s experiment setup. I am now trying to have a larger scope and incorporate different type of data:, for instance:
– Buridan’s experiment done with a different tracker
– Walking honeybee tracking in a rectangular arena, with a rewarded target
– Animal (flies/bees) walking on a ball, using open- or closed-loop experiment setup
– trajectory data obtained from the pysolo software (flies)
– larval crawling data
I want to include an automatic depository of the data in a database. Automatic entries in Figshare is for instance possible. (see older posts). My problem is to find a way to treat the data such that:
1. the raw data is uploaded
2. all data is uploaded also if we use only the centroid displacement (in some data file the head position is also given)
3. the data can be reused and data obtained in different lab, animal, setup can be compared. (data should be organized such that it can be searched and queried).
4. probably other elements that I do not think of….
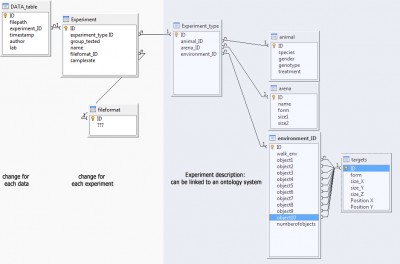
My main problem: I have nearly no experience in data management/design, ontology or semantic web. Here is a first draft of a database structure that I have thought of. Every feedback would be welcome:
Category: buridan, open science, Pysolo, Spontaneous Behavior

Here’s a book that I intend to read before the meeting in February and that may contain some answers:
https://www.sciencedirect.com/science/bookseries/00747742/103/supp/C